Concurrent loss of LKB1 and KEAP1 enhances SHMT-mediated antioxidant defence in KRAS-mutant lung cancer
- PMID: 38877143
- PMCID: PMC11809267
- DOI: 10.1038/s42255-024-01066-z
Concurrent loss of LKB1 and KEAP1 enhances SHMT-mediated antioxidant defence in KRAS-mutant lung cancer
Abstract
Non-small-cell lung cancer (NSCLC) with concurrent mutations in KRAS and the tumour suppressor LKB1 (KL NSCLC) is refractory to most therapies and has one of the worst predicted outcomes. Here we describe a KL-induced metabolic vulnerability associated with serine-glycine-one-carbon (SGOC) metabolism. Using RNA-seq and metabolomics data from human NSCLC, we uncovered that LKB1 loss enhanced SGOC metabolism via serine hydroxymethyltransferase (SHMT). LKB1 loss, in collaboration with KEAP1 loss, activated SHMT through inactivation of the salt-induced kinase (SIK)-NRF2 axis and satisfied the increased demand for one-carbon units necessary for antioxidant defence. Chemical and genetic SHMT suppression increased cellular sensitivity to oxidative stress and cell death. Further, the SHMT inhibitor enhanced the in vivo therapeutic efficacy of paclitaxel (first-line NSCLC therapy inducing oxidative stress) in KEAP1-mutant KL tumours. The data reveal how this highly aggressive molecular subtype of NSCLC fulfills their metabolic requirements and provides insight into therapeutic strategies.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
W.R.E. and K.O. are employees of the Barer Institute, which is engaged in the clinical development of SHMT inhibitors. H.W. is a consultant for the following pharmaceutical companies: Genentech, Denali Therapeutics, Alector, Surrozen, Cleave Therapeutics, ORIC Pharmaceuticals, Barer Institute, Vincerx, Chinook and Cresenta. J.K. (UTSW) is on the scientific advisory board for Sanofi and is a consultant for Pulmatrix. The other authors declare no competing interests.
Figures







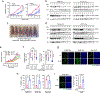
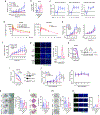








References
-
- Lunt SY & Vander Heiden MG Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol 27, 441–464 (2011). - PubMed
-
- Osthus RC et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem 275, 21797–21800 (2000). - PubMed
MeSH terms
Substances
Grants and funding
- IA-828202/American Lung Association (Lung Association)
- R37 CA285640/CA/NCI NIH HHS/United States
- K22 CA226676/CA/NCI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- RSG-21-153-01-CCB/American Cancer Society (American Cancer Society, Inc.)
- P50CA070907/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P01HL160469/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 1R01CA258684/U.S. Department of Health & Human Services | NIH | NCI | Division of Cancer Epidemiology and Genetics, National Cancer Institute (National Cancer Institute Division of Cancer Epidemiology and Genetics)
- R01 CA258684/CA/NCI NIH HHS/United States
- P30CA177558/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P01 HL160469/HL/NHLBI NIH HHS/United States
- R01 CA196851/CA/NCI NIH HHS/United States
- R00 CA215307/CA/NCI NIH HHS/United States
- P30 CA177558/CA/NCI NIH HHS/United States
- 1K22CA226676-01A1/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R00 CA237724/CA/NCI NIH HHS/United States
- R00CA237724/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P50 CA070907/CA/NCI NIH HHS/United States
- 1R37CA285640-01A1/U.S. Department of Health & Human Services | NIH | NCI | Division of Cancer Epidemiology and Genetics, National Cancer Institute (National Cancer Institute Division of Cancer Epidemiology and Genetics)
- 1R01CA196851/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R00CA215307/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P50 CA196530/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

