ABIN1 is a negative regulator of effector functions in cytotoxic T cells
- PMID: 38877170
- PMCID: PMC11315980
- DOI: 10.1038/s44319-024-00179-6
ABIN1 is a negative regulator of effector functions in cytotoxic T cells
Abstract
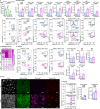
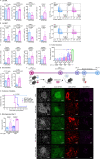
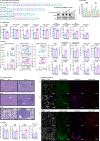
T cells are pivotal in the adaptive immune defense, necessitating a delicate balance between robust response against infections and self-tolerance. Their activation involves intricate cross-talk among signaling pathways triggered by the T-cell antigen receptors (TCR) and co-stimulatory or inhibitory receptors. The molecular regulation of these complex signaling networks is still incompletely understood. Here, we identify the adaptor protein ABIN1 as a component of the signaling complexes of GITR and OX40 co-stimulation receptors. T cells lacking ABIN1 are hyper-responsive ex vivo, exhibit enhanced responses to cognate infections, and superior ability to induce experimental autoimmune diabetes in mice. ABIN1 negatively regulates p38 kinase activation and late NF-κB target genes. P38 is at least partially responsible for the upregulation of the key effector proteins IFNG and GZMB in ABIN1-deficient T cells after TCR stimulation. Our findings reveal the intricate role of ABIN1 in T-cell regulation.
Keywords: ABIN1; Antigen Receptor; Co-stimulation; T Cells; p38.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









Similar articles
-
Dual-specificity phosphatase 14 (DUSP14/MKP6) negatively regulates TCR signaling by inhibiting TAB1 activation.J Immunol. 2014 Feb 15;192(4):1547-57. doi: 10.4049/jimmunol.1300989. Epub 2014 Jan 8. J Immunol. 2014. PMID: 24403530
-
Negative regulation of interleukin-2 and p38 mitogen-activated protein kinase during T-cell activation by the adaptor ALX.Mol Cell Biol. 2006 Aug;26(16):6005-15. doi: 10.1128/MCB.02067-05. Mol Cell Biol. 2006. PMID: 16880512 Free PMC article.
-
A20-binding inhibitor of NF-κB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein β activation and protects from inflammatory disease.Proc Natl Acad Sci U S A. 2011 Nov 1;108(44):E998-1006. doi: 10.1073/pnas.1106232108. Epub 2011 Oct 19. Proc Natl Acad Sci U S A. 2011. PMID: 22011580 Free PMC article.
-
The role of OX40-mediated co-stimulation in T-cell activation and survival.Crit Rev Immunol. 2009;29(3):187-201. doi: 10.1615/critrevimmunol.v29.i3.10. Crit Rev Immunol. 2009. PMID: 19538134 Free PMC article. Review.
-
Signaling triggered by glucocorticoid-induced tumor necrosis factor receptor family-related gene: regulation at the interface between regulatory T cells and immune effector cells.Front Biosci. 2006 May 1;11:1448-65. doi: 10.2741/1895. Front Biosci. 2006. PMID: 16368528 Review.
References
-
- Connolly KA, Kuchroo M, Venkat A, Khatun A, Wang J, William I, Hornick NI, Fitzgerald BL, Damo M, Kasmani MY et al (2021) A reservoir of stem-like CD8(+) T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci Immunol 6:eabg7836 10.1126/sciimmunol.abg7836 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 22-18046S/Grantová Agentura České Republiky (GAČR)
- LX22NPO5103/European Union - Next Generation EU
- IG 4420/European Molecular Biology Organization (EMBO)
- PRIMUS/20/MED/003/Univerzita Karlova v Praze (UK)
- 984120/Grantová Agentura, Univerzita Karlova (GA UK)
- 274323/Grantová Agentura, Univerzita Karlova (GA UK)
- 802878/EC | ERC | HORIZON EUROPE European Research Council (ERC)
- OP RDI CZ.1.05/2.1.00/19.0395/Ministerstvo Školství, Mládeže a Tělovýchovy (MŠMT)
- LM2023036/Ministerstvo Školství, Mládeže a Tělovýchovy (MŠMT)
- OP RDI BIOCEV CZ.1.05/1.1.00/02.0109/Ministerstvo Školství, Mládeže a Tělovýchovy (MŠMT)
- LM2023050/Ministerstvo Školství, Mládeže a Tělovýchovy (MŠMT)
- RVO - 68378050-KAV-NPUI/Ministerstvo Školství, Mládeže a Tělovýchovy (MŠMT)
- RVO 68378050/Ústav molekulární genetiky, AVČR core funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

