Genetic and environmental interactions contribute to immune variation in rewilded mice
- PMID: 38877178
- PMCID: PMC11224019
- DOI: 10.1038/s41590-024-01862-5
Genetic and environmental interactions contribute to immune variation in rewilded mice
Abstract
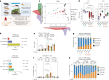
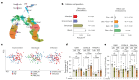
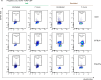
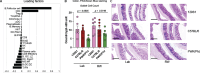
The relative and synergistic contributions of genetics and environment to interindividual immune response variation remain unclear, despite implications in evolutionary biology and medicine. Here we quantify interactive effects of genotype and environment on immune traits by investigating C57BL/6, 129S1 and PWK/PhJ inbred mice, rewilded in an outdoor enclosure and infected with the parasite Trichuris muris. Whereas cellular composition was shaped by interactions between genotype and environment, cytokine response heterogeneity including IFNγ concentrations was primarily driven by genotype with consequence on worm burden. In addition, we show that other traits, such as expression of CD44, were explained mostly by genetics on T cells, whereas expression of CD44 on B cells was explained more by environment across all strains. Notably, genetic differences under laboratory conditions were decreased following rewilding. These results indicate that nonheritable influences interact with genetic factors to shape immune variation and parasite burden.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
K.C. has received research funding from Pfizer, Takeda, Pacific Biosciences, Genentech and Abbvie, and P.L. has received research funding from Pfizer. K.C. has consulted for or received an honorarium from Puretech Health, Genentech and Abbvie. K.C. is an inventor on US patent 10,722,600 and provisional patents 62/935,035 and 63/157,225. S.B.K. acknowledges funding from Micreos and KymeraTx in the past 3 years. P.L. and O.O. are federal employees. The other authors declare no competing interests.
Figures














Update of
-
Genetic and Environmental interactions contribute to immune variation in rewilded mice.bioRxiv [Preprint]. 2023 May 2:2023.03.17.533121. doi: 10.1101/2023.03.17.533121. bioRxiv. 2023. Update in: Nat Immunol. 2024 Jul;25(7):1270-1282. doi: 10.1038/s41590-024-01862-5. PMID: 36993484 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

