The crystal structure of methanogen McrD, a methyl-coenzyme M reductase-associated protein
- PMID: 38877345
- PMCID: PMC11301259
- DOI: 10.1002/2211-5463.13848
The crystal structure of methanogen McrD, a methyl-coenzyme M reductase-associated protein
Abstract
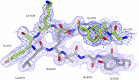
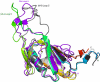
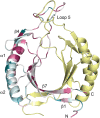
Methyl-coenzyme M reductase (MCR) is a multi-subunit (α2β2γ2) enzyme responsible for methane formation via its unique F430 cofactor. The genes responsible for producing MCR (mcrA, mcrB and mcrG) are typically colocated with two other highly conserved genes mcrC and mcrD. We present here the high-resolution crystal structure for McrD from a human gut methanogen Methanomassiliicoccus luminyensis strain B10. The structure reveals that McrD comprises a ferredoxin-like domain assembled into an α + β barrel-like dimer with conformational flexibility exhibited by a functional loop. The description of the M. luminyensis McrD crystal structure contributes to our understanding of this key conserved methanogen protein typically responsible for promoting MCR activity and the production of methane, a greenhouse gas.
Keywords: Methanomassiliicoccus luminyensis; McrD; ferredoxin‐like; methanogen; methyl‐coenzyme M reductase.
© 2024 The Author(s). FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
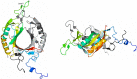
References
-
- Shima S, Warkentin E, Thauer RK and Ermler U (2002) Structure and function of enzymes involved in the methanogenic pathway utilizing carbon dioxide and molecular hydrogen. J Biosci Bioeng 93, 519–530. - PubMed
-
- Kaster AK, Goenrich M, Seedorf H, Liesegang H, Wollherr A, Gottschalk G and Thauer RK (2011) More than 200 genes required for methane formation from H2 and CO2 and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus . Archaea 2011, 973848. - PMC - PubMed
-
- Scheller S, Goenrich M, Boecher R, Thauer RK and Jaun B (2010) The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608. - PubMed
-
- Ermler U, Grabarse W, Shima S, Goubeaud M and Thauer RK (1997) Crystal structure of methyl‐coenzyme M reductase: the key enzyme of biological methane formation. Science 278, 1457–1462. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

