Revealing the Arabidopsis AtGRP7 mRNA binding proteome by specific enhanced RNA interactome capture
- PMID: 38877390
- PMCID: PMC11177498
- DOI: 10.1186/s12870-024-05249-4
Revealing the Arabidopsis AtGRP7 mRNA binding proteome by specific enhanced RNA interactome capture
Abstract
Background: The interaction of proteins with RNA in the cell is crucial to orchestrate all steps of RNA processing. RNA interactome capture (RIC) techniques have been implemented to catalogue RNA- binding proteins in the cell. In RIC, RNA-protein complexes are stabilized by UV crosslinking in vivo. Polyadenylated RNAs and associated proteins are pulled down from cell lysates using oligo(dT) beads and the RNA-binding proteome is identified by quantitative mass spectrometry. However, insights into the RNA-binding proteome of a single RNA that would yield mechanistic information on how RNA expression patterns are orchestrated, are scarce.
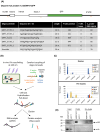
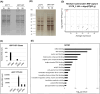
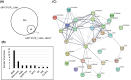
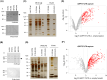
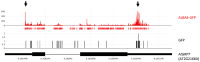
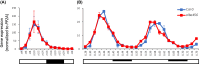
Results: Here, we explored RIC in Arabidopsis to identify proteins interacting with a single mRNA, using the circadian clock-regulated Arabidopsis thaliana GLYCINE-RICH RNA-BINDING PROTEIN 7 (AtGRP7) transcript, one of the most abundant transcripts in Arabidopsis, as a showcase. Seedlings were treated with UV light to covalently crosslink RNA and proteins. The AtGRP7 transcript was captured from cell lysates with antisense oligonucleotides directed against the 5'untranslated region (UTR). The efficiency of RNA capture was greatly improved by using locked nucleic acid (LNA)/DNA oligonucleotides, as done in the enhanced RIC protocol. Furthermore, performing a tandem capture with two rounds of pulldown with the 5'UTR oligonucleotide increased the yield. In total, we identified 356 proteins enriched relative to a pulldown from atgrp7 mutant plants. These were benchmarked against proteins pulled down from nuclear lysates by AtGRP7 in vitro transcripts immobilized on beads. Among the proteins validated by in vitro interaction we found the family of Acetylation Lowers Binding Affinity (ALBA) proteins. Interaction of ALBA4 with the AtGRP7 RNA was independently validated via individual-nucleotide resolution crosslinking and immunoprecipitation (iCLIP). The expression of the AtGRP7 transcript in an alba loss-of-function mutant was slightly changed compared to wild-type, demonstrating the functional relevance of the interaction.
Conclusion: We adapted specific RNA interactome capture with LNA/DNA oligonucleotides for use in plants using AtGRP7 as a showcase. We anticipate that with further optimization and up scaling the protocol should be applicable for less abundant transcripts.
Keywords: Arabidopsis; Enhanced RNA interactome capture; LNA oligonucleotides; iCLIP.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

