Analysis of a new phage, KZag1, infecting biofilm of Klebsiella pneumoniae: genome sequence and characterization
- PMID: 38877452
- PMCID: PMC11179266
- DOI: 10.1186/s12866-024-03355-9
Analysis of a new phage, KZag1, infecting biofilm of Klebsiella pneumoniae: genome sequence and characterization
Abstract
Background: This study investigates the effectiveness of the bacteriophage KZag1 against drug-resistant Klebsiella pneumoniae, aiming to assess its potential as a therapeutic agent. The novelty lies in the characterization of KZag1, a Myovirus with specific efficacy against multidrug-resistant K. pneumoniae strains. This highlights the significance of exploring alternative strategies, particularly phage therapy, in addressing biofilm-associated infections.
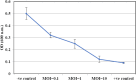
Methods: KZag1, characterized by a typical Myovirus structure with a 75 ± 5 nm diameter icosahedral head and a 15 ± 5 nm short tail, was evaluated in experimental trials against 15 strains of K. pneumoniae. The infection cycle duration was determined to be 50 min, resulting in an estimated burst size of approximately 83 plaque-forming units per colony-forming unit (PFU/CFU). Stability assessments were conducted within a pH range of 4 to 12 and temperatures ranging from 45°C to 60°C. Biofilm biomass reduction was observed, particularly at a multiplicity of infection (MOI) of 10.
Results: KZag1 demonstrated infection efficacy against 12 out of 15 tested K. pneumoniae strains. The phage exhibited stability across a broad pH range and at elevated temperatures. Notably, treatment with KZag1 significantly reduced K. pneumoniae biofilm biomass, emphasizing its potential in combating biofilm formation. Genomic analysis revealed a complete genome of 157,089 base pairs with a GC content of 46.38%, encompassing 203 open reading frames (ORFs) and a cysteine-specific tRNA sequence. Comparison with phage GP4 highlighted similarities, with KZag1 having a longer genome by approximately 4829 base pairs and a higher GC content by approximately 0.93%. Phylogenetic analysis classified KZag1 within the Myoviridae family.
Conclusion: The efficacy of KZag1 against K. pneumoniae biofilm suggests its potential as a therapeutic candidate, especially for drug-resistant infections. Further clinical research is warranted to explore its synergy with other treatments, elucidate genomic traits, compare with Myoviridae phages, and understand its host interactions. These findings underscore the promising role of KZag1 in addressing drug-resistant bacterial infections.
Keywords: K. pneumoniae; And Antibiotic resistance; Bacteriophage KZag1; Biofilm reduction; Genomic analysis; Phage Therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Phage Therapy: A Promising Treatment Strategy against Infections Caused by Multidrug-resistant Klebsiella pneumoniae.Curr Pharm Des. 2025;31(13):1007-1019. doi: 10.2174/0113816128343976241117183624. Curr Pharm Des. 2025. PMID: 39757682 Review.
-
Isolation, Identification, and Biological Characterization of Phage vB_KpnM_KpVB3 Targeting Carbapenem-Resistant Klebsiella pneumoniae ST11.J Glob Antimicrob Resist. 2024 Jun;37:179-184. doi: 10.1016/j.jgar.2024.03.007. Epub 2024 Mar 30. J Glob Antimicrob Resist. 2024. PMID: 38561142
-
Isolation and characterization of a novel phage targeting Klebsiella pneumoniae K2 capsular type.Braz J Microbiol. 2025 Sep;56(3):1887-1900. doi: 10.1007/s42770-025-01707-9. Epub 2025 Jun 6. Braz J Microbiol. 2025. PMID: 40481305
-
Isolation, characterization, therapeutic potency, and genomic analysis of a novel bacteriophage vB_KshKPC-M against carbapenemase-producing Klebsiella pneumoniae strains (CRKP) isolated from Ventilator-associated pneumoniae (VAP) infection of COVID-19 patients.Ann Clin Microbiol Antimicrob. 2023 Feb 24;22(1):18. doi: 10.1186/s12941-023-00567-1. Ann Clin Microbiol Antimicrob. 2023. PMID: 36829156 Free PMC article.
-
The potential use of bacteriophages as antibacterial agents against Klebsiella pneumoniae.Virol J. 2024 Aug 19;21(1):191. doi: 10.1186/s12985-024-02450-7. Virol J. 2024. PMID: 39160541 Free PMC article. Review.
Cited by
-
Analysis of a novel phage as a promising biological agent targeting multidrug resistant Klebsiella pneumoniae.AMB Express. 2025 Mar 5;15(1):37. doi: 10.1186/s13568-025-01846-0. AMB Express. 2025. PMID: 40044971 Free PMC article.
-
Phage Therapy: A Promising Treatment Strategy against Infections Caused by Multidrug-resistant Klebsiella pneumoniae.Curr Pharm Des. 2025;31(13):1007-1019. doi: 10.2174/0113816128343976241117183624. Curr Pharm Des. 2025. PMID: 39757682 Review.
References
-
- Farzana K, Rizvi M, Al-Shahrani AM. Klebsiella pneumoniae: An Emerging Pathogen of Healthcare Setting in Riyadh. Saudi Arabia J Pure Appl Microbiol. 2019;13(1):49–54.
-
- Qureshi ZA. Infections due to multidrug-resistant Gram-negative bacteria among hospitalized individuals in an Indian teaching hospital. Indian J Med Microbiol. 2016;34:45–51.
-
- Chhibber S, Kaur S, Kumari S. Therapeutic potential of bacteriophage in treating Klebsiella pneumoniae B5055-mediated lobar pneumonia in mice. J Med Microbiol. 2013;62(Pt 8):1143–1152. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

