Beyond traditional translation: ncRNA derived peptides as modulators of tumor behaviors
- PMID: 38877495
- PMCID: PMC11177406
- DOI: 10.1186/s12929-024-01047-0
Beyond traditional translation: ncRNA derived peptides as modulators of tumor behaviors
Abstract
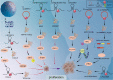
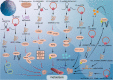
Within the intricate tapestry of molecular research, noncoding RNAs (ncRNAs) were historically overshadowed by a pervasive presumption of their inability to encode proteins or peptides. However, groundbreaking revelations have challenged this notion, unveiling select ncRNAs that surprisingly encode peptides specifically those nearing a succinct 100 amino acids. At the forefront of this epiphany stand lncRNAs and circRNAs, distinctively characterized by their embedded small open reading frames (sORFs). Increasing evidence has revealed different functions and mechanisms of peptides/proteins encoded by ncRNAs in cancer, including promotion or inhibition of cancer cell proliferation, cellular metabolism (glucose metabolism and lipid metabolism), and promotion or concerted metastasis of cancer cells. The discoveries not only accentuate the depth of ncRNA functionality but also open novel avenues for oncological research and therapeutic innovations. The main difficulties in the study of these ncRNA-derived peptides hinge crucially on precise peptide detection and sORFs identification. Here, we illuminate cutting-edge methodologies, essential instrumentation, and dedicated databases tailored for unearthing sORFs and peptides. In addition, we also conclude the potential of clinical applications in cancer therapy.
Keywords: Cancer; Peptides; Protein; circRNAs; lncRNAs; ncRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Noncoding RNA-encoded peptides in cancer: biological functions, posttranslational modifications and therapeutic potential.J Hematol Oncol. 2025 Feb 19;18(1):20. doi: 10.1186/s13045-025-01671-9. J Hematol Oncol. 2025. PMID: 39972384 Free PMC article. Review.
-
Cancer-related micropeptides encoded by ncRNAs: Promising drug targets and prognostic biomarkers.Cancer Lett. 2022 Oct 28;547:215723. doi: 10.1016/j.canlet.2022.215723. Epub 2022 May 7. Cancer Lett. 2022. PMID: 35533953 Review.
-
ncRNA-Encoded Peptides or Proteins and Cancer.Mol Ther. 2019 Oct 2;27(10):1718-1725. doi: 10.1016/j.ymthe.2019.09.001. Epub 2019 Sep 6. Mol Ther. 2019. PMID: 31526596 Free PMC article. Review.
-
Subcellular localization and relevant mechanisms of human cancer-related micropeptides.FASEB J. 2023 Dec;37(12):e23270. doi: 10.1096/fj.202301019RR. FASEB J. 2023. PMID: 37994683 Review.
-
Identification and analysis of short open reading frames (sORFs) in the initially annotated noncoding RNA LINC00493 from human cells.J Biochem. 2021 Apr 29;169(4):421-434. doi: 10.1093/jb/mvaa143. J Biochem. 2021. PMID: 33386847
Cited by
-
The Role and Function of Non-Coding RNAs in Cholangiocarcinoma Invasiveness.Biomedicines. 2025 Jun 3;13(6):1369. doi: 10.3390/biomedicines13061369. Biomedicines. 2025. PMID: 40564088 Free PMC article. Review.
-
lncRNAs: the unexpected link between protein synthesis and cancer adaptation.Mol Cancer. 2025 Jan 31;24(1):38. doi: 10.1186/s12943-025-02236-7. Mol Cancer. 2025. PMID: 39891197 Free PMC article. Review.
-
Unlocking the secrets of the immunopeptidome: MHC molecules, ncRNA peptides, and vesicles in immune response.Front Immunol. 2025 Jan 29;16:1540431. doi: 10.3389/fimmu.2025.1540431. eCollection 2025. Front Immunol. 2025. PMID: 39944685 Free PMC article. Review.
-
Noncoding RNA-encoded peptides in cancer: biological functions, posttranslational modifications and therapeutic potential.J Hematol Oncol. 2025 Feb 19;18(1):20. doi: 10.1186/s13045-025-01671-9. J Hematol Oncol. 2025. PMID: 39972384 Free PMC article. Review.
-
Beyond the Transcript: Translating Non-Coding RNAs and Their Impact on Cellular Regulation.Cancers (Basel). 2025 May 3;17(9):1555. doi: 10.3390/cancers17091555. Cancers (Basel). 2025. PMID: 40361481 Free PMC article. Review.
References
-
- Consortium EP. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Program N.C.S., Baylor College of Medicine Human Genome Sequencing C., Washington University Genome Sequencing C., Broad I., Children's Hospital Oakland Research I. Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. - DOI - PMC - PubMed
-
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. - DOI - PMC - PubMed
-
- Bo H, Fan L, Li J, Liu Z, Zhang S, Shi L, Guo C, Li X, Liao Q, Zhang W, Zhou M, Xiang B, Li X, Li G, Xiong W, Zeng Z, Xiong F, Gong Z. High Expression of lncRNA AFAP1-AS1 promotes the progression of colon cancer and predicts poor prognosis. J Cancer. 2018;9(24):4677–4683. doi: 10.7150/jca.26461. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

