The C. elegans Myc-family of transcription factors coordinate a dynamic adaptive response to dietary restriction
- PMID: 38878153
- PMCID: PMC11336136
- DOI: 10.1007/s11357-024-01197-x
The C. elegans Myc-family of transcription factors coordinate a dynamic adaptive response to dietary restriction
Abstract
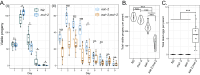
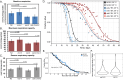
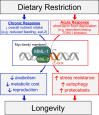
Dietary restriction (DR), the process of decreasing overall food consumption over an extended period of time, has been shown to increase longevity across evolutionarily diverse species and delay the onset of age-associated diseases in humans. In Caenorhabditis elegans, the Myc-family transcription factors (TFs) MXL-2 (Mlx) and MML-1 (MondoA/ChREBP), which function as obligate heterodimers, and PHA-4 (orthologous to FOXA) are both necessary for the full physiological benefits of DR. However, the adaptive transcriptional response to DR and the role of MML-1::MXL-2 and PHA-4 remains elusive. We identified the transcriptional signature of C. elegans DR, using the eat-2 genetic model, and demonstrate broad changes in metabolic gene expression in eat-2 DR animals, which requires both mxl-2 and pha-4. While the requirement for these factors in DR gene expression overlaps, we found many of the DR genes exhibit an opposing change in relative gene expression in eat-2;mxl-2 animals compared to wild-type, which was not observed in eat-2 animals with pha-4 loss. Surprisingly, we discovered more than 2000 genes synthetically dysregulated in eat-2;mxl-2, out of which the promoters of down-regulated genes were substantially enriched for PQM-1 and ELT-1/3 GATA TF binding motifs. We further show functional deficiencies of the mxl-2 loss in DR outside of lifespan, as eat-2;mxl-2 animals exhibit substantially smaller brood sizes and lay a proportion of dead eggs, indicating that MML-1::MXL-2 has a role in maintaining the balance between resource allocation to the soma and to reproduction under conditions of chronic food scarcity. While eat-2 animals do not show a significantly different metabolic rate compared to wild-type, we also find that loss of mxl-2 in DR does not affect the rate of oxygen consumption in young animals. The gene expression signature of eat-2 mutant animals is consistent with optimization of energy utilization and resource allocation, rather than induction of canonical gene expression changes associated with acute metabolic stress, such as induction of autophagy after TORC1 inhibition. Consistently, eat-2 animals are not substantially resistant to stress, providing further support to the idea that chronic DR may benefit healthspan and lifespan through efficient use of limited resources rather than broad upregulation of stress responses, and also indicates that MML-1::MXL-2 and PHA-4 may have distinct roles in promotion of benefits in response to different pro-longevity stimuli.
Keywords: Dietary restriction; Gene expression; Myc-family; Transcription factors.
© 2024. The Author(s), under exclusive licence to American Aging Association.
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
The C. elegans Myc-family of transcription factors coordinate a dynamic adaptive response to dietary restriction.bioRxiv [Preprint]. 2023 Nov 22:2023.11.22.568222. doi: 10.1101/2023.11.22.568222. bioRxiv. 2023. Update in: Geroscience. 2024 Oct;46(5):4827-4854. doi: 10.1007/s11357-024-01197-x. PMID: 38045350 Free PMC article. Updated. Preprint.
References
-
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

