Conservation of C4BP-binding sequence patterns in Streptococcus pyogenes M and Enn proteins
- PMID: 38879009
- PMCID: PMC11292367
- DOI: 10.1016/j.jbc.2024.107478
Conservation of C4BP-binding sequence patterns in Streptococcus pyogenes M and Enn proteins
Abstract
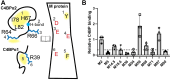
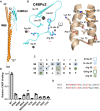
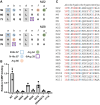
Antigenically sequence variable M proteins of the major bacterial pathogen Streptococcus pyogenes (Strep A) are responsible for recruiting human C4b-binding protein (C4BP) to the bacterial surface, which enables Strep A to evade destruction by the immune system. The most sequence divergent portion of M proteins, the hypervariable region (HVR), is responsible for binding C4BP. Structural evidence points to the conservation of two C4BP-binding sequence patterns (M2 and M22) in the HVR of numerous M proteins, with this conservation applicable to vaccine immunogen design. These two patterns, however, only partially explain C4BP binding by Strep A. Here, we identified several M proteins that lack these patterns but still bind C4BP and determined the structures of two, M68 and M87 HVRs, in complex with a C4BP fragment. Mutagenesis of these M proteins led to the identification of amino acids that are crucial for C4BP binding, enabling formulation of new C4BP-binding patterns. Mutagenesis was also carried out on M2 and M22 proteins to refine or generate experimentally grounded C4BP-binding patterns. The M22 pattern was the most prevalent among M proteins, followed by the M87 and M2 patterns, while the M68 pattern was rare. These patterns, except for M68, were also evident in numerous M-like Enn proteins. Binding of C4BP via these patterns to Enn proteins was verified. We conclude that C4BP-binding patterns occur frequently in Strep A strains of differing M types, being present in their M or Enn proteins, or frequently both, providing further impetus for their use as vaccine immunogens.
Keywords: C4BP; M protein; Streptococcus pyogenes; cross-reactivity; immunogen.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Update of
-
Conservation of C4BP-binding Sequence Patterns in Streptococcus pyogenes M and Enn Proteins.bioRxiv [Preprint]. 2024 Apr 22:2024.04.22.590534. doi: 10.1101/2024.04.22.590534. bioRxiv. 2024. Update in: J Biol Chem. 2024 Jul;300(7):107478. doi: 10.1016/j.jbc.2024.107478. PMID: 38712057 Free PMC article. Updated. Preprint.
References
-
- Nitsche-Schmitz D.P., Chhatwal G.S. Host-pathogen interactions in streptococcal immune sequelae. Curr. Top. Microbiol. Immunol. 2013;368:155–171. - PubMed
-
- Carapetis J.R., Steer A.C., Mulholland E.K., Weber M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005;5:685–694. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

