The origin of esterase activity of Parkinson's disease causative factor DJ-1 implied by evolutionary trace analysis of its prokaryotic homolog HchA
- PMID: 38879013
- PMCID: PMC11301059
- DOI: 10.1016/j.jbc.2024.107476
The origin of esterase activity of Parkinson's disease causative factor DJ-1 implied by evolutionary trace analysis of its prokaryotic homolog HchA
Abstract
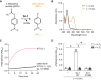
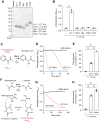
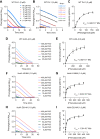
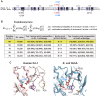
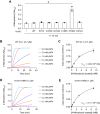
DJ-1, a causative gene for hereditary recessive Parkinsonism, is evolutionarily conserved across eukaryotes and prokaryotes. Structural analyses of DJ-1 and its homologs suggested the 106th Cys is a nucleophilic cysteine functioning as the catalytic center of hydratase or hydrolase activity. Indeed, DJ-1 and its homologs can convert highly electrophilic α-oxoaldehydes such as methylglyoxal into α-hydroxy acids as hydratase in vitro, and oxidation-dependent ester hydrolase (esterase) activity has also been reported for DJ-1. The mechanism underlying such plural activities, however, has not been fully characterized. To address this knowledge gap, we conducted a series of biochemical assays assessing the enzymatic activity of DJ-1 and its homologs. We found no evidence for esterase activity in any of the Escherichia coli DJ-1 homologs. Furthermore, contrary to previous reports, we found that oxidation inactivated rather than facilitated DJ-1 esterase activity. The E. coli DJ-1 homolog HchA possesses phenylglyoxalase and methylglyoxalase activities but lacks esterase activity. Since evolutionary trace analysis identified the 186th H as a candidate residue involved in functional differentiation between HchA and DJ-1, we focused on H186 of HchA and found that an esterase activity was acquired by H186A mutation. Introduction of reverse mutations into the equivalent position in DJ-1 (A107H) selectively eliminated its esterase activity without compromising α-oxoaldehyde hydratase activity. The obtained results suggest that differences in the amino acid sequences near the active site contributed to acquisition of esterase activity in vitro and provide an important clue to the origin and significance of DJ-1 esterase activity.
Keywords: DJ-1; PARK7; Parkinson disease (PD); catalytic triad; esterase; hydratase; oxo-aldehyde; prokaryote.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interests with the contents of this article.
Figures


Similar articles
-
Novel Redox-Dependent Esterase Activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson's Disease.Int J Mol Sci. 2016 Aug 22;17(8):1346. doi: 10.3390/ijms17081346. Int J Mol Sci. 2016. PMID: 27556455 Free PMC article.
-
Structural Biology of the DJ-1 Superfamily.Adv Exp Med Biol. 2017;1037:5-24. doi: 10.1007/978-981-10-6583-5_2. Adv Exp Med Biol. 2017. PMID: 29147900
-
Parkinson's disease-related DJ-1 functions in thiol quality control against aldehyde attack in vitro.Sci Rep. 2017 Oct 9;7(1):12816. doi: 10.1038/s41598-017-13146-0. Sci Rep. 2017. PMID: 28993701 Free PMC article.
-
DJ-1/PARK7: A New Therapeutic Target for Neurodegenerative Disorders.Biol Pharm Bull. 2017;40(5):548-552. doi: 10.1248/bpb.b16-01006. Biol Pharm Bull. 2017. PMID: 28458339 Review.
-
Recent findings on the physiological function of DJ-1: Beyond Parkinson's disease.Neurobiol Dis. 2017 Dec;108:65-72. doi: 10.1016/j.nbd.2017.08.005. Epub 2017 Aug 18. Neurobiol Dis. 2017. PMID: 28823929 Review.
Cited by
-
The reaction mechanism for glycolysis side product degradation by Parkinson's disease-linked DJ-1.J Cell Biol. 2025 Aug 4;224(8):e202411078. doi: 10.1083/jcb.202411078. Epub 2025 Jun 4. J Cell Biol. 2025. PMID: 40464736 Free PMC article.
References
-
- Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. - PubMed
-
- Kahle P.J., Waak J., Gasser T. DJ-1 and prevention of oxidative stress in Parkinson's disease and other age-related disorders. Free Radic. Biol. Med. 2009;47:1354–1361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

