Integrated analysis of transcriptomics and metabolomics of garden asparagus (Asparagus officinalis L.) under drought stress
- PMID: 38879466
- PMCID: PMC11179350
- DOI: 10.1186/s12870-024-05286-z
Integrated analysis of transcriptomics and metabolomics of garden asparagus (Asparagus officinalis L.) under drought stress
Abstract
Background: Drought is a leading environmental factor affecting plant growth. To explore the drought tolerance mechanism of asparagus, this study analyzed the responses of two asparagus varieties, namely, 'Jilv3' (drought tolerant) and 'Pacific Early' (drought sensitive), to drought stress using metabolomics and transcriptomics.
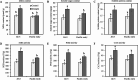
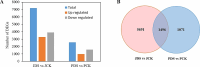
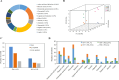
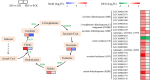
Results: In total, 2,567 and 7,187 differentially expressed genes (DEGs) were identified in 'Pacific Early' and 'Jilv3', respectively, by comparing the transcriptome expression patterns between the normal watering treatment and the drought stress treatment. These DEGs were significantly enriched in the amino acid biosynthesis, carbon metabolism, phenylpropanoid biosynthesis, and plant hormone signal transduction pathways. In 'Jilv3', DEGs were also enriched in the following energy metabolism-related pathways: citrate cycle (TCA cycle), glycolysis/gluconeogenesis, and pyruvate metabolism. This study also identified 112 and 254 differentially accumulated metabolites (DAMs) in 'Pacific Early' and 'Jilv3' under drought stress compared with normal watering, respectively. The amino acid, flavonoid, organic acid, and soluble sugar contents were more significantly enhanced in 'Jilv3' than in 'Pacific Early'. According to the metabolome and transcriptome analysis, in 'Jilv3', the energy supply of the TCA cycle was improved, and flavonoid biosynthesis increased. As a result, its adaptability to drought stress improved.
Conclusions: These findings help to better reveal the molecular mechanism underlying how asparagus responds to drought stress and improve researchers' ability to screen drought-tolerant asparagus varieties as well as breed new varieties.
Keywords: Asparagus; Drought stress; Metabolic pathways; Metabolome; Transcriptome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Transcriptomic and Metabolomic Profiling of Root Tissue in Drought-Tolerant and Drought-Susceptible Wheat Genotypes in Response to Water Stress.Int J Mol Sci. 2024 Sep 27;25(19):10430. doi: 10.3390/ijms251910430. Int J Mol Sci. 2024. PMID: 39408761 Free PMC article.
-
Comparative transcriptome analysis of the garden asparagus (Asparagus officinalis L.) reveals the molecular mechanism for growth with arbuscular mycorrhizal fungi under salinity stress.Plant Physiol Biochem. 2019 Aug;141:20-29. doi: 10.1016/j.plaphy.2019.05.013. Epub 2019 May 15. Plant Physiol Biochem. 2019. PMID: 31125808
-
Transcriptome sequencing and metabolome analysis reveal the molecular mechanism of Salvia miltiorrhiza in response to drought stress.BMC Plant Biol. 2024 May 23;24(1):446. doi: 10.1186/s12870-024-05006-7. BMC Plant Biol. 2024. PMID: 38778268 Free PMC article.
-
Transcriptome expression profiles reveal response mechanisms to drought and drought-stress mitigation mechanisms by exogenous glycine betaine in maize.Biotechnol Lett. 2022 Mar;44(3):367-386. doi: 10.1007/s10529-022-03221-6. Epub 2022 Mar 16. Biotechnol Lett. 2022. PMID: 35294695 Review.
-
Exploring Plant Resilience Through Secondary Metabolite Profiling: Advances in Stress Response and Crop Improvement.Plant Cell Environ. 2025 Jul;48(7):4823-4837. doi: 10.1111/pce.15473. Epub 2025 Mar 17. Plant Cell Environ. 2025. PMID: 40091600 Review.
Cited by
-
Physiological and metabolomic analyses reveal the mechanism by which exogenous spermine improves drought resistance in alfalfa leaves (Medicago sativa L.).Front Plant Sci. 2024 Oct 9;15:1466493. doi: 10.3389/fpls.2024.1466493. eCollection 2024. Front Plant Sci. 2024. PMID: 39445141 Free PMC article.
References
-
- Yang A, Akhtar S, Li L, Fu Q, et al. Biochar mitigates combined effects of drought and salinity stress in quinoa. Agronomy. 2020;10(6):912. doi: 10.3390/agronomy10060912. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

