Catechin-Induced changes in PODXL, DNMTs, and miRNA expression in Nalm6 cells: an integrated in silico and in vitro approach
- PMID: 38879474
- PMCID: PMC11179370
- DOI: 10.1186/s12906-024-04521-2
Catechin-Induced changes in PODXL, DNMTs, and miRNA expression in Nalm6 cells: an integrated in silico and in vitro approach
Abstract
Background: This study explored the impact of predicted miRNAs on DNA methyltransferases (DNMTs) and the PODXL gene in Nalm6 cells, revealing the significance of these miRNAs in acute lymphocytic leukemia (ALL).
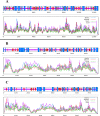
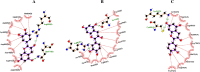
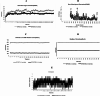
Methods: A comprehensive approach was adopted, integrating bioinformatic analyses encompassing protein structure prediction, molecular docking, dynamics, and ADMET profiling, in conjunction with evaluations of gene and miRNA expression patterns. This methodology was employed to elucidate the therapeutic potential of catechin compounds in modulating the activity of DNA methyltransferases (DNMTs) and the PODXL gene.
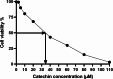
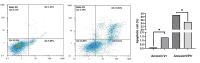
Results: The findings from our investigation indicate that catechins possess the capability to inhibit DNMT enzymes. This inhibitory effect is associated with the upregulation of microRNAs miR-200c and miR-548 and a concurrent downregulation of PODXL gene expression. These molecular interactions culminate in an augmented apoptotic response within ALL (Nalm6) cells.
Conclusion: The study posits that catechins may represent a viable therapeutic avenue for inducing apoptosis in ALL cells. This is achieved through the modulation of epigenetic mechanisms and alterations in gene expression profiles, highlighting the potential of catechins as agents for cancer therapy.
Keywords: ALL; And catechin; Docking; MD simulation; Methyltransferase; PODXL; miRNAs.
© 2024. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Figures
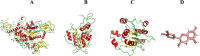



Similar articles
-
6-Gingerol modulates miRNAs and PODXL gene expression via methyltransferase enzymes in NB4 cells: an in silico and in vitro study.Sci Rep. 2024 Aug 7;14(1):18356. doi: 10.1038/s41598-024-68069-4. Sci Rep. 2024. PMID: 39112503 Free PMC article.
-
Mechanisms of DNA methyltransferase-inhibitor interactions: Procyanidin B2 shows new promise for therapeutic intervention of cancer.Chem Biol Interact. 2015 May 25;233:122-38. doi: 10.1016/j.cbi.2015.03.022. Epub 2015 Mar 31. Chem Biol Interact. 2015. PMID: 25839702
-
[Effect of MiR-142-3p Targeting HOXA5 on Proliferation, Cycle Arrest and Apoptosis of Acute B Lymphocytic Leukemia Cells].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2021 Aug;29(4):1085-1092. doi: 10.19746/j.cnki.issn.1009-2137.2021.04.011. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2021. PMID: 34362486 Chinese.
-
Discovery and development of DNA methyltransferase inhibitors using in silico approaches.Drug Discov Today. 2015 May;20(5):569-77. doi: 10.1016/j.drudis.2014.12.007. Epub 2014 Dec 16. Drug Discov Today. 2015. PMID: 25526932 Review.
-
The epigenetic feedback loop between DNA methylation and microRNAs in fibrotic disease with an emphasis on DNA methyltransferases.Cell Signal. 2013 Sep;25(9):1870-6. doi: 10.1016/j.cellsig.2013.05.013. Epub 2013 May 22. Cell Signal. 2013. PMID: 23707521 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

