CPMI: comprehensive neighborhood-based perturbed mutual information for identifying critical states of complex biological processes
- PMID: 38879513
- PMCID: PMC11180411
- DOI: 10.1186/s12859-024-05836-0
CPMI: comprehensive neighborhood-based perturbed mutual information for identifying critical states of complex biological processes
Abstract
Background: There exists a critical transition or tipping point during the complex biological process. Such critical transition is usually accompanied by the catastrophic consequences. Therefore, hunting for the tipping point or critical state is of significant importance to prevent or delay the occurrence of catastrophic consequences. However, predicting critical state based on the high-dimensional small sample data is a difficult problem, especially for single-cell expression data.
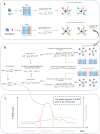
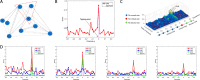
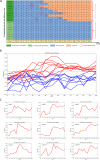
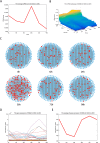
Results: In this study, we propose the comprehensive neighbourhood-based perturbed mutual information (CPMI) method to detect the critical states of complex biological processes. The CPMI method takes into account the relationship between genes and neighbours, so as to reduce the noise and enhance the robustness. This method is applied to a simulated dataset and six real datasets, including an influenza dataset, two single-cell expression datasets and three bulk datasets. The method can not only successfully detect the tipping points, but also identify their dynamic network biomarkers (DNBs). In addition, the discovery of transcription factors (TFs) which can regulate DNB genes and nondifferential 'dark genes' validates the effectiveness of our method. The numerical simulation verifies that the CPMI method is robust under different noise strengths and is superior to the existing methods on identifying the critical states.
Conclusions: In conclusion, we propose a robust computational method, i.e., CPMI, which is applicable in both the bulk and single cell datasets. The CPMI method holds great potential in providing the early warning signals for complex biological processes and enabling early disease diagnosis.
Keywords: Dark genes; Dynamic network biomarker (DNB); Perturbed neighbourhood mutual information (PMI); Tipping point; Transcription factors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

