Title and abstract screening for literature reviews using large language models: an exploratory study in the biomedical domain
- PMID: 38879534
- PMCID: PMC11180407
- DOI: 10.1186/s13643-024-02575-4
Title and abstract screening for literature reviews using large language models: an exploratory study in the biomedical domain
Abstract
Background: Systematically screening published literature to determine the relevant publications to synthesize in a review is a time-consuming and difficult task. Large language models (LLMs) are an emerging technology with promising capabilities for the automation of language-related tasks that may be useful for such a purpose.
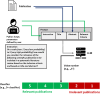
Methods: LLMs were used as part of an automated system to evaluate the relevance of publications to a certain topic based on defined criteria and based on the title and abstract of each publication. A Python script was created to generate structured prompts consisting of text strings for instruction, title, abstract, and relevant criteria to be provided to an LLM. The relevance of a publication was evaluated by the LLM on a Likert scale (low relevance to high relevance). By specifying a threshold, different classifiers for inclusion/exclusion of publications could then be defined. The approach was used with four different openly available LLMs on ten published data sets of biomedical literature reviews and on a newly human-created data set for a hypothetical new systematic literature review.
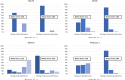
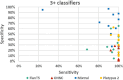
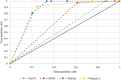
Results: The performance of the classifiers varied depending on the LLM being used and on the data set analyzed. Regarding sensitivity/specificity, the classifiers yielded 94.48%/31.78% for the FlanT5 model, 97.58%/19.12% for the OpenHermes-NeuralChat model, 81.93%/75.19% for the Mixtral model and 97.58%/38.34% for the Platypus 2 model on the ten published data sets. The same classifiers yielded 100% sensitivity at a specificity of 12.58%, 4.54%, 62.47%, and 24.74% on the newly created data set. Changing the standard settings of the approach (minor adaption of instruction prompt and/or changing the range of the Likert scale from 1-5 to 1-10) had a considerable impact on the performance.
Conclusions: LLMs can be used to evaluate the relevance of scientific publications to a certain review topic and classifiers based on such an approach show some promising results. To date, little is known about how well such systems would perform if used prospectively when conducting systematic literature reviews and what further implications this might have. However, it is likely that in the future researchers will increasingly use LLMs for evaluating and classifying scientific publications.
Keywords: Biomedicine; Large language models; Natural language processing; Systematic literature review; Title and abstract screening.
© 2024. The Author(s).
Conflict of interest statement
NC is a technical lead for the SmartOncology© project and medical advisor for Wemedoo AG, Steinhausen AG, Switzerland. The authors declare that they have no other competing interests.
Figures
References
-
- van de Schoot R, de Bruin J, Schram R, Zahedi P, de Boer J, Weijdema F, et al. An open source machine learning framework for efficient and transparent systematic reviews. Nat Mach Intell. 2021;3(2):125–133. doi: 10.1038/s42256-020-00287-7. - DOI

