A tripartite organelle platform links growth factor receptor signaling to mitochondrial metabolism
- PMID: 38879572
- PMCID: PMC11180189
- DOI: 10.1038/s41467-024-49543-z
A tripartite organelle platform links growth factor receptor signaling to mitochondrial metabolism
Abstract
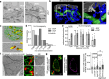
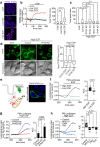
One open question in the biology of growth factor receptors is how a quantitative input (i.e., ligand concentration) is decoded by the cell to produce specific response(s). Here, we show that an EGFR endocytic mechanism, non-clathrin endocytosis (NCE), which is activated only at high ligand concentrations and targets receptor to degradation, requires a tripartite organelle platform involving the plasma membrane (PM), endoplasmic reticulum (ER) and mitochondria. At these contact sites, EGFR-dependent, ER-generated Ca2+ oscillations are sensed by mitochondria, leading to increased metabolism and ATP production. Locally released ATP is required for cortical actin remodeling and EGFR-NCE vesicle fission. The same biochemical circuitry is also needed for an effector function of EGFR, i.e., collective motility. The multiorganelle signaling platform herein described mediates direct communication between EGFR signaling and mitochondrial metabolism, and is predicted to have a broad impact on cell physiology as it is activated by another growth factor receptor, HGFR/MET.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- AIRC IG 24415/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- AIRC IG 18988/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- AIRC IG 23060/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- MCO 10000/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- 20-0094/AICR_/Worldwide Cancer Research/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

