Fusaric acid mediates the assembly of disease-suppressive rhizosphere microbiota via induced shifts in plant root exudates
- PMID: 38879580
- PMCID: PMC11180119
- DOI: 10.1038/s41467-024-49218-9
Fusaric acid mediates the assembly of disease-suppressive rhizosphere microbiota via induced shifts in plant root exudates
Abstract
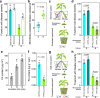
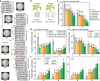
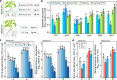
The plant health status is determined by the interplay of plant-pathogen-microbiota in the rhizosphere. Here, we investigate this tripartite system focusing on the pathogen Fusarium oxysporum f. sp. lycopersici (FOL) and tomato plants as a model system. First, we explore differences in tomato genotype resistance to FOL potentially associated with the differential recruitment of plant-protective rhizosphere taxa. Second, we show the production of fusaric acid by FOL to trigger systemic changes in the rhizosphere microbiota. Specifically, we show this molecule to have opposite effects on the recruitment of rhizosphere disease-suppressive taxa in the resistant and susceptible genotypes. Last, we elucidate that FOL and fusaric acid induce changes in the tomato root exudation with direct effects on the recruitment of specific disease-suppressive taxa. Our study unravels a mechanism mediating plant rhizosphere assembly and disease suppression by integrating plant physiological responses to microbial-mediated mechanisms in the rhizosphere.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Volatile-mediated interspecific plant interaction promotes root colonization by beneficial bacteria via induced shifts in root exudation.Microbiome. 2024 Oct 21;12(1):207. doi: 10.1186/s40168-024-01914-w. Microbiome. 2024. PMID: 39428455 Free PMC article.
-
Fusarium oxysporum f. sp. lycopersici C2H2 transcription factor FolCzf1 is required for conidiation, fusaric acid production, and early host infection.Curr Genet. 2019 Jun;65(3):773-783. doi: 10.1007/s00294-019-00931-9. Epub 2019 Jan 10. Curr Genet. 2019. PMID: 30631890
-
Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling.Proc Natl Acad Sci U S A. 2020 Feb 18;117(7):3874-3883. doi: 10.1073/pnas.1912130117. Epub 2020 Feb 3. Proc Natl Acad Sci U S A. 2020. PMID: 32015118 Free PMC article.
-
Root exudates: from plant to rhizosphere and beyond.Plant Cell Rep. 2020 Jan;39(1):3-17. doi: 10.1007/s00299-019-02447-5. Epub 2019 Jul 25. Plant Cell Rep. 2020. PMID: 31346716 Review.
-
Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes.Curr Opin Microbiol. 2019 Jun;49:73-82. doi: 10.1016/j.mib.2019.10.003. Epub 2019 Nov 13. Curr Opin Microbiol. 2019. PMID: 31731229 Review.
Cited by
-
Total Deoxynivalenol Contamination of Wheat Products and Coarse Grains in Shanghai, China: Occurrence and Health Risk Assessment.Foods. 2024 Oct 23;13(21):3373. doi: 10.3390/foods13213373. Foods. 2024. PMID: 39517157 Free PMC article.
-
Fusaric acid-mediated S-glutathionylation of MaAKT1 channel confers the virulence of Foc TR4 to banana.PLoS Pathog. 2025 Apr 9;21(4):e1013066. doi: 10.1371/journal.ppat.1013066. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40203070 Free PMC article.
-
Metabolite-mediated responses of phyllosphere microbiota to powdery mildew infection in resistant and susceptible black currant cultivars.Hortic Res. 2025 Mar 25;12(7):uhaf092. doi: 10.1093/hr/uhaf092. eCollection 2025 Jul. Hortic Res. 2025. PMID: 40371437 Free PMC article.
-
Exploring plant-microbe interactions in adapting to abiotic stress under climate change: a review.Front Plant Sci. 2024 Nov 15;15:1482739. doi: 10.3389/fpls.2024.1482739. eCollection 2024. Front Plant Sci. 2024. PMID: 39619840 Free PMC article. Review.
-
Genome-Wide Identification of the Rehmannia glutinosa miRNA Family and Exploration of Their Expression Characteristics Caused by the Replant Disease Formation-Related Principal Factor.Genes (Basel). 2024 Sep 23;15(9):1239. doi: 10.3390/genes15091239. Genes (Basel). 2024. PMID: 39336830 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

