Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate
- PMID: 38879612
- PMCID: PMC11180146
- DOI: 10.1038/s41467-024-48802-3
Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate
Abstract
Bacteroidales (syn. Bacteroidetes) are prominent members of the human gastrointestinal ecosystem mainly due to their efficient glycan-degrading machinery, organized into gene clusters known as polysaccharide utilization loci (PULs). A single PUL was reported for catabolism of high-mannose (HM) N-glycan glyco-polypeptides in the gut symbiont Bacteroides thetaiotaomicron, encoding a surface endo-β-N-acetylglucosaminidase (ENGase), BT3987. Here, we discover an ENGase from the GH18 family in B. thetaiotaomicron, BT1285, encoded in a distinct PUL with its own repertoire of proteins for catabolism of the same HM N-glycan substrate as that of BT3987. We employ X-ray crystallography, electron microscopy, mass spectrometry-based activity measurements, alanine scanning mutagenesis and a broad range of biophysical methods to comprehensively define the molecular mechanism by which BT1285 recognizes and hydrolyzes HM N-glycans, revealing that the stabilities and activities of BT1285 and BT3987 were optimal in markedly different conditions. BT1285 exhibits significantly higher affinity and faster hydrolysis of poorly accessible HM N-glycans than does BT3987. We also find that two HM-processing endoglycosidases from the human gut-resident Alistipes finegoldii display condition-specific functional properties. Altogether, our data suggest that human gut microbes employ evolutionary strategies to express distinct ENGases in order to optimally metabolize the same N-glycan substrate in the gastroinstestinal tract.
© 2024. The Author(s).
Conflict of interest statement
Based on the development of an artificial lectin, BT1285i, with high specificity and high affinity for oligomannose
Figures

 GlcNAc,
GlcNAc,  Man,
Man,  Fuc,
Fuc,  Gal,
Gal,  Neu5A. Source data are provided as a Source Data file.
Neu5A. Source data are provided as a Source Data file.
 GlcNAc,
GlcNAc,  Man. β-1-4 bond cleaved by ENGases is indicated with scissors. d Close inspection of substrate binding site of BT1285 in complex with Man9GlcNAc2. Residues used in the alanine scanning assays are represented as sticks. e Hydrolytic activity of BT1285 and mutants against HM-IgG1, as determined by LC-MS analysis, normalized to BT1285 wt. Data are presented as mean values±SD. Assays were run in technical triplicates (n = 3). f Close inspection of key residues involved in the Mannoses recognition and binding in BT1285. Source data are provided as a Source Data file.
Man. β-1-4 bond cleaved by ENGases is indicated with scissors. d Close inspection of substrate binding site of BT1285 in complex with Man9GlcNAc2. Residues used in the alanine scanning assays are represented as sticks. e Hydrolytic activity of BT1285 and mutants against HM-IgG1, as determined by LC-MS analysis, normalized to BT1285 wt. Data are presented as mean values±SD. Assays were run in technical triplicates (n = 3). f Close inspection of key residues involved in the Mannoses recognition and binding in BT1285. Source data are provided as a Source Data file.
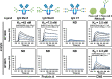
 GlcNAc,
GlcNAc,  Man,
Man,  Fuc,
Fuc,  Gal. Source data are provided as a Source Data file.
Gal. Source data are provided as a Source Data file.
 GlcNAc,
GlcNAc,  Man. Source data are provided as a Source Data file.
Man. Source data are provided as a Source Data file.

 GlcNAc,
GlcNAc,  Man,
Man,  Fuc,
Fuc,  Gal,
Gal,  Neu5A. Source data are provided as a Source Data file.
Neu5A. Source data are provided as a Source Data file.References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

