Lactylation of NAT10 promotes N4-acetylcytidine modification on tRNASer-CGA-1-1 to boost oncogenic DNA virus KSHV reactivation
- PMID: 38879723
- PMCID: PMC11445560
- DOI: 10.1038/s41418-024-01327-0
Lactylation of NAT10 promotes N4-acetylcytidine modification on tRNASer-CGA-1-1 to boost oncogenic DNA virus KSHV reactivation
Abstract
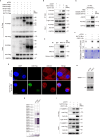
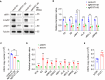
N4-acetylcytidine (ac4C), a conserved but recently rediscovered RNA modification on tRNAs, rRNAs and mRNAs, is catalyzed by N-acetyltransferase 10 (NAT10). Lysine acylation is a ubiquitous protein modification that controls protein functions. Our latest study demonstrates a NAT10-dependent ac4C modification, which occurs on the polyadenylated nuclear RNA (PAN) encoded by oncogenic DNA virus Kaposi's sarcoma-associated herpesvirus (KSHV), can induce KSHV reactivation from latency and activate inflammasome. However, it remains unclear whether a novel lysine acylation occurs in NAT10 during KSHV reactivation and how this acylation of NAT10 regulates tRNAs ac4C modification. Here, we showed that NAT10 was lactylated by α-tubulin acetyltransferase 1 (ATAT1), as a writer at the critical domain, to exert RNA acetyltransferase function and thus increase the ac4C level of tRNASer-CGA-1-1. Mutagenesis at the ac4C site in tRNASer-CGA-1-1 inhibited its ac4C modifications, translation efficiency of viral lytic genes, and virion production. Mechanistically, KSHV PAN orchestrated NAT10 and ATAT1 to enhance NAT10 lactylation, resulting in tRNASer-CGA-1-1 ac4C modification, eventually boosting KSHV reactivation. Our findings reveal a novel post-translational modification in NAT10, as well as expand the understanding about tRNA-related ac4C modification during KSHV replication, which may be exploited to design therapeutic strategies for KSHV-related diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
NAT10-dependent N4-acetylcytidine modification mediates PAN RNA stability, KSHV reactivation, and IFI16-related inflammasome activation.Nat Commun. 2023 Oct 10;14(1):6327. doi: 10.1038/s41467-023-42135-3. Nat Commun. 2023. PMID: 37816771 Free PMC article.
-
NAT10 and N4-acetylcytidine restrain R-loop levels and related inflammatory responses.Sci Adv. 2025 Mar 28;11(13):eads6144. doi: 10.1126/sciadv.ads6144. Epub 2025 Mar 26. Sci Adv. 2025. PMID: 40138394 Free PMC article.
-
The Expression and Nuclear Retention Element of Polyadenylated Nuclear RNA Is Not Required for Productive Lytic Replication of Kaposi's Sarcoma-Associated Herpesvirus.J Virol. 2021 Jun 10;95(13):e0009621. doi: 10.1128/JVI.00096-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853955 Free PMC article.
-
Emerging Role of NAT10 as ac4C Writer in Inflammatory Diseases: Mechanisms and Therapeutic Applications.Curr Drug Targets. 2025;26(4):282-294. doi: 10.2174/0113894501346709241202110834. Curr Drug Targets. 2025. PMID: 39633518 Review.
-
Regulation of KSHV Latency and Lytic Reactivation.Viruses. 2020 Sep 17;12(9):1034. doi: 10.3390/v12091034. Viruses. 2020. PMID: 32957532 Free PMC article. Review.
Cited by
-
Metabolism Meets Translation: Dietary and Metabolic Influences on tRNA Modifications and Codon Biased Translation.Wiley Interdiscip Rev RNA. 2025 Mar-Apr;16(2):e70011. doi: 10.1002/wrna.70011. Wiley Interdiscip Rev RNA. 2025. PMID: 40119534 Free PMC article. Review.
-
The lactylation-macrophage interplay: implications for gastrointestinal disease therapeutics.Front Immunol. 2025 Jul 9;16:1608115. doi: 10.3389/fimmu.2025.1608115. eCollection 2025. Front Immunol. 2025. PMID: 40703527 Free PMC article. Review.
-
Lactylation-regulated biomolecular condensates: metabolic control of phase separation in physiology and disease.Cell Commun Signal. 2025 May 25;23(1):239. doi: 10.1186/s12964-025-02244-6. Cell Commun Signal. 2025. PMID: 40414883 Free PMC article. Review.
-
How lactate and lactylation shape the immunity system in atherosclerosis (Review).Int J Mol Med. 2025 Oct;56(4):163. doi: 10.3892/ijmm.2025.5604. Epub 2025 Aug 1. Int J Mol Med. 2025. PMID: 40747658 Free PMC article. Review.
-
Non-histone lactylation: unveiling its functional significance.Front Cell Dev Biol. 2025 Jan 24;13:1535611. doi: 10.3389/fcell.2025.1535611. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 39925738 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 82241067/National Natural Science Foundation of China (National Science Foundation of China)
- 82072275/National Natural Science Foundation of China (National Science Foundation of China)
- 32170151/National Natural Science Foundation of China (National Science Foundation of China)
- 81871642/National Natural Science Foundation of China (National Science Foundation of China)
- 31800148/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

