Regulation of primary cilia disassembly through HUWE1-mediated TTBK2 degradation plays a crucial role in cerebellar development and medulloblastoma growth
- PMID: 38879724
- PMCID: PMC11445238
- DOI: 10.1038/s41418-024-01325-2
Regulation of primary cilia disassembly through HUWE1-mediated TTBK2 degradation plays a crucial role in cerebellar development and medulloblastoma growth
Abstract
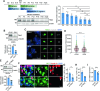
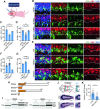
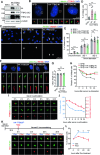
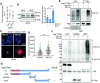
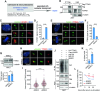
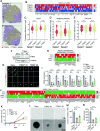
Development of the cerebellum requires precise regulation of granule neuron progenitor (GNP) proliferation. Although it is known that primary cilia are necessary to support GNP proliferation, the exact molecular mechanism governing primary cilia dynamics within GNPs remains elusive. Here, we establish the pivotal roles for the centrosomal kinase TTBK2 (Tau tubulin kinase-2) and the E3 ubiquitin ligase HUWE1 in GNP proliferation. We show that TTBK2 is highly expressed in proliferating GNPs under Sonic Hedgehog (SHH) signaling, coinciding with active GNP proliferation and the presence of primary cilia. TTBK2 stabilizes primary cilia by inhibiting their disassembly, thereby promoting GNP proliferation in response to SHH. Mechanistically, we identify HUWE1 as a novel centrosomal E3 ligase that facilitates primary cilia disassembly by targeting TTBK2 degradation. Disassembly of primary cilia serves as a trigger for GNP differentiation, allowing their migration from the external granule layer (EGL) of the cerebellum to the internal granule layer (IGL) for subsequent maturation. Moreover, we have established a link between TTBK2 and SHH-type medulloblastoma (SHH-MB), a tumor characterized by uncontrolled GNP proliferation. TTBK2 depletion inhibits SHH-MB proliferation, indicating that TTBK2 may be a potential therapeutic target for this cancer type. In summary, our findings reveal the mechanism governing cerebellar development and highlight a potential anti-cancer strategy for SHH-MB.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Butts T, Green MJ, Wingate RJ. Development of the cerebellum: simple steps to make a ‘little brain. Development. 2014;141:4031–41. - PubMed
-
- Adamaszek M, D’Agata F, Ferrucci R, Habas C, Keulen S, Kirkby KC, et al. Consensus paper: cerebellum and emotion. Cerebellum. 2017;16:552–76. - PubMed
-
- Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–8. - PubMed
-
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic hedgehog. Neuron. 1999;22:103–14. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

