Evolutionary Dynamics of Accelerated Antiviral Resistance Development in Hypermutator Herpesvirus
- PMID: 38879872
- PMCID: PMC11226790
- DOI: 10.1093/molbev/msae119
Evolutionary Dynamics of Accelerated Antiviral Resistance Development in Hypermutator Herpesvirus
Abstract
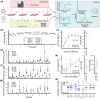
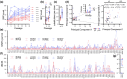
Antiviral therapy is constantly challenged by the emergence of resistant pathogens. At the same time, experimental approaches to understand and predict resistance are limited by long periods required for evolutionary processes. Here, we present a herpes simplex virus 1 mutant with impaired proofreading capacity and consequently elevated mutation rates. Comparing this hypermutator to parental wild type virus, we study the evolution of antiviral drug resistance in vitro. We model resistance development and elucidate underlying genetic changes against three antiviral substances. Our analyzes reveal no principle difference in the evolutionary behavior of both viruses, adaptive processes are overall similar, however significantly accelerated for the hypermutator. We conclude that hypermutator viruses are useful for modeling adaptation to antiviral therapy. They offer the benefit of expedited adaptation without introducing apparent bias and can therefore serve as an accelerator to predict natural evolution.
Keywords: HSV-1; accelerated; antiviral resistance; experimental evolution; hypermutation.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures





Similar articles
-
Fast-forwarding evolution-Accelerated adaptation in a proofreading-deficient hypermutator herpesvirus.Virus Evol. 2022 Oct 14;8(2):veac099. doi: 10.1093/ve/veac099. eCollection 2022. Virus Evol. 2022. PMID: 36405341 Free PMC article.
-
A mutation in the DNA polymerase accessory factor of herpes simplex virus 1 restores viral DNA replication in the presence of raltegravir.J Virol. 2014 Oct;88(19):11121-9. doi: 10.1128/JVI.01540-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008933 Free PMC article.
-
Genotypic characterization of UL23 thymidine kinase and UL30 DNA polymerase of clinical isolates of herpes simplex virus: natural polymorphism and mutations associated with resistance to antivirals.Antimicrob Agents Chemother. 2010 Nov;54(11):4833-42. doi: 10.1128/AAC.00669-10. Epub 2010 Aug 23. Antimicrob Agents Chemother. 2010. PMID: 20733037 Free PMC article.
-
Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management.Antimicrob Agents Chemother. 2011 Feb;55(2):459-72. doi: 10.1128/AAC.00615-10. Epub 2010 Nov 15. Antimicrob Agents Chemother. 2011. PMID: 21078929 Free PMC article. Review.
-
Herpes simplex virus helicase-primase inhibitors: recent findings from the study of drug resistance mutations.Antivir Chem Chemother. 2008;19(1):1-6. doi: 10.1177/095632020801900101. Antivir Chem Chemother. 2008. PMID: 18610552 Review.
Cited by
-
Protective Mechanisms of Vaginal Lactobacilli against Sexually Transmitted Viral Infections.Int J Mol Sci. 2024 Aug 23;25(17):9168. doi: 10.3390/ijms25179168. Int J Mol Sci. 2024. PMID: 39273118 Free PMC article. Review.
References
-
- Bennett L, Melchers B, Proppe B. Curta: a general-purpose high-performance computer at ZEDAT, Freie Universität Berlin. 2020. 10.17169/refubium-26754. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

