Profiling Proteins and Phosphorylation Sites During T Cell Activation Using an Integrated Thermal Shift Assay
- PMID: 38880243
- PMCID: PMC11298636
- DOI: 10.1016/j.mcpro.2024.100801
Profiling Proteins and Phosphorylation Sites During T Cell Activation Using an Integrated Thermal Shift Assay
Abstract
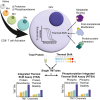
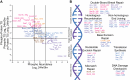
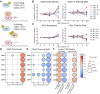
T cell activation is a complex biological process of naive cells maturing into effector cells. Proteomic and phospho-proteomic approaches have provided critical insights into this process, yet it is not always clear how changes in individual proteins or phosphorylation sites have functional significance. Here, we developed the Phosphorylation Integrated Thermal Shift Assay (PITSA) that combines the measurement of protein or phosphorylation site abundance and thermal stability into a single tandem mass tags experiment and apply this method to study T cell activation. We quantified the abundance and thermal stability of over 7500 proteins and 5000 phosphorylation sites and identified significant differences in chromatin-related, TCR signaling, DNA repair, and proliferative phosphoproteins. PITSA may be applied to a wide range of biological contexts to generate hypotheses as to which proteins or phosphorylation sites are functionally regulated in a given system as well as the mechanisms by which this regulation may occur.
Keywords: CD8((+)) T cell activation; DNA repair; cyclin-dependent kinase signaling; phosphoproteomics; proteome thermal stability; proteomics.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interests The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: S. P. G. is a member of the scientific advisory boards of Cell Signaling Technologies and ThermoFisher Scientific. L. C. C. is a founder and member of the board of directors of Agios Pharmaceuticals and is a founder and receives research support from Petra Pharmaceuticals; is listed as an inventor on a patent (WO2019232403A1, Weill Cornell Medicine) for combination therapy for PI3K-associated disease or disorder, and the identification of therapeutic interventions to improve response to PI3K inhibitors for cancer treatment; is a co-founder and shareholder in Faeth Therapeutics; has equity in and consults for Cell Signaling Technologies, Volastra, Larkspur and 1 Base Pharmaceuticals; and consults for Loxo-Lilly. J. L. J has received consulting fees from Scorpion Therapeutics and Volastra Therapeutics. T. M. Y.-B. is a co-founder of DeStroke.
Figures




Similar articles
-
Mass Spectrometry-Based Phosphoproteomics and Systems Biology: Approaches to Study T Lymphocyte Activation and Exhaustion.J Mol Biol. 2021 Dec 3;433(24):167318. doi: 10.1016/j.jmb.2021.167318. Epub 2021 Oct 24. J Mol Biol. 2021. PMID: 34687714 Review.
-
LymphoAtlas: a dynamic and integrated phosphoproteomic resource of TCR signaling in primary T cells reveals ITSN2 as a regulator of effector functions.Mol Syst Biol. 2020 Jul;16(7):e9524. doi: 10.15252/msb.20209524. Mol Syst Biol. 2020. PMID: 32618424 Free PMC article.
-
Quantitative phosphoproteomic analysis reveals a role for serine and threonine kinases in the cytoskeletal reorganization in early T cell receptor activation in human primary T cells.Mol Cell Proteomics. 2012 May;11(5):171-86. doi: 10.1074/mcp.M112.017863. Epub 2012 Apr 12. Mol Cell Proteomics. 2012. PMID: 22499768 Free PMC article.
-
Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis.Sci Signal. 2010 Jan 12;3(104):ra3. doi: 10.1126/scisignal.2000475. Sci Signal. 2010. PMID: 20068231
-
Phosphoproteomic analysis: an emerging role in deciphering cellular signaling in human embryonic stem cells and their differentiated derivatives.Stem Cell Rev Rep. 2012 Mar;8(1):16-31. doi: 10.1007/s12015-011-9317-8. Stem Cell Rev Rep. 2012. PMID: 22009073 Free PMC article. Review.
References
-
- Ron-Harel N., Sharpe A.H., Haigis M.C. Mitochondrial metabolism in T cell activation and senescence: a mini-review. Gerontology. 2015;61:131–138. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

