Evaluating therapeutic potential of AYUSH-64 constituents against omicron variant of SARS-CoV-2 using ensemble docking and simulations
- PMID: 38881558
- PMCID: PMC11179622
- DOI: 10.1016/j.crstbi.2024.100151
Evaluating therapeutic potential of AYUSH-64 constituents against omicron variant of SARS-CoV-2 using ensemble docking and simulations
Abstract
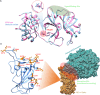
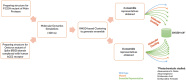
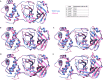
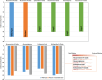
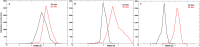
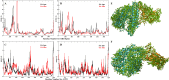
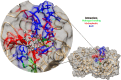
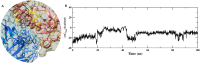
The COVID-19 pandemic in the later phase showed the presence of the B.1.1.529 variant of the SARS-CoV-2 designated as Omicron. AYUSH-64 a poly herbal drug developed by Central Council for Research in Ayurvedic Sciences (CCRAS) has been recommended by Ministry of Ayush in asymptomatic, mild to moderate COVID-19 patients. One of the earlier, in-silico study has shown the binding of the constituents of AYUSH-64 to the main protease (Mpro) of the SARS-CoV-2. This study enlisted four phytochemicals of AYUSH-64, which were found to have significant binding with the Mpro. In continuation to the same, the current study proposes to understand the binding of these four phytochemicals to main protease (Mpro) and receptor binding domain (RBD) of spike protein of the Omicron variant. An enhanced molecular docking methodology, namely, ensemble docking has been used to find the most efficiently binding phytochemical. Using molecular dynamics (MD) simulations and clustering approach it was observed that the Mpro and RBD Spike of Omicron variant of SARS-CoV-2 in complex with human ACE2 tends to attain 4 and 8 conformational respectively. Based on the docking studies, the best binding phytochemical of the AYUSH-64, akummicine N-oxide was selected for MD simulations. MD simulations of akummicine N-oxide bound to omicron variant of Mpro and RBD Spike-ACE complex was performed. The conformational, interaction and binding energy analysis suggested that the akummicine N-oxide binds well with Mpro and RBD Spike-ACE2 complex. The interaction between RBD Spike and ACE2 was observed to weaken in the presence of akummicine N-oxide. Hence, it can be inferred that, these phytochemicals from AYUSH-64 formulation may have the potential to act against the Omicron variant of SARS-CoV-2.
Keywords: Ayurveda; Ayush64; Ensemble docking; MD simulations; Phytochemicals.
© 2024 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures








References
-
- Allerton C.M.N., Arcari J.T., Aschenbrenner L.M., Avery M., Bechle B.M., Behzadi M.A., Boras B., Buzon L.M., Cardin R.D., Catlin N.R., Carlo A.A., Coffman K.J., Dantonio A., Di L., Eng H., Farley K.A., Ferre R.A., Gernhardt S.S., Gibson S.A., Greasley S.E., Greenfield S.R., Hurst B.L., Kalgutkar A.S., Kimoto E., Lanyon L.F., Lovett G.H., Lian Y., Liu W., Martínez Alsina L.A., Noell S., Obach R.S., Owen D.R., Patel N.C., Rai D.K., Reese M.R., Rothan H.A., Sakata S., Sammons M.F., Sathish J.G., Sharma R., Steppan C.M., Tuttle J.B., Verhoest P.R., Wei L., Yang Q., Yurgelonis I., Zhu Y. A second-generation oral SARS-CoV-2 main protease inhibitor clinical candidate for the treatment of COVID-19. J. Med. Chem. 2024;30 doi: 10.1021/acs.jmedchem.3c02469. Epub ahead of print. PMID: 38687966. - DOI - PMC - PubMed
-
- AMBER 2020. 2020.
-
- Cha-Silva A.S., Gavaghan M.B., Bergroth T., Alexander-Parrish R., Yang J., Draica F., Patel J., Garner D.A., Stanford R.H., Meier G., McLaughlin J.M., Nguyen J.L. Effectiveness of nirmatrelvir-ritonavir for the prevention of COVID-19-related hospitalization and mortality: a systematic literature review. Am. J. Therapeut. 2024;31(3):e246–e257. doi: 10.1097/MJT.0000000000001744. Epub 2024 Apr 29. PMID: 38691664; PMCID: PMC11060058. - DOI - PMC - PubMed
-
- Chopra A., Tillu G., Chuadhary K., Reddy G., Srivastava A., Lakdawala M., Gode D., Reddy H., Tamboli S., Saluja M., Sarmukaddam S., Gundeti M., Raut A.K., Rao B.C.S., Yadav B., Srikanth N., Patwardhan B. Co-administration of AYUSH 64 as an adjunct to standard of care in mild and moderate COVID-19: a randomized, controlled, multicentric clinical trial. PLoS One. 2023;18(3) doi: 10.1371/JOURNAL.PONE.0282688. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

