Overexpression of arginase gene CAR1 renders yeast Saccharomyces cerevisiae acetic acid tolerance
- PMID: 38882181
- PMCID: PMC11178985
- DOI: 10.1016/j.synbio.2024.05.013
Overexpression of arginase gene CAR1 renders yeast Saccharomyces cerevisiae acetic acid tolerance
Abstract
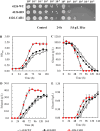
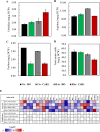
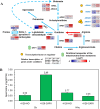
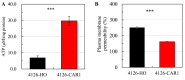
Acetic acid is a common inhibitor present in lignocellulose hydrolysate, which inhibits the ethanol production by yeast strains. Therefore, the cellulosic ethanol industry requires yeast strains that can tolerate acetic acid stress. Here we demonstrate that overexpressing a yeast native arginase-encoding gene, CAR1, renders Saccharomyces cerevisiae acetic acid tolerance. Specifically, ethanol yield increased by 27.3% in the CAR1-overexpressing strain compared to the control strain under 5.0 g/L acetic acid stress. The global intracellular amino acid level and compositions were further analyzed, and we found that CAR1 overexpression reduced the total amino acid content in response to acetic acid stress. Moreover, the CAR1 overexpressing strain showed increased ATP level and improved cell membrane integrity. Notably, we demonstrated that the effect of CAR1 overexpression was independent of the spermidine and proline metabolism, which indicates novel mechanisms for enhancing yeast stress tolerance. Our studies also suggest that CAR1 is a novel genetic element to be used in synthetic biology of yeast for efficient production of fuel ethanol.
Keywords: Acetic acid tolerance; Amino acid metabolism; Arginase; CAR1; Ethanol production; Saccharomyces cerevisiae.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Protective Effects of Arginine on Saccharomyces cerevisiae Against Ethanol Stress.Sci Rep. 2016 Aug 10;6:31311. doi: 10.1038/srep31311. Sci Rep. 2016. PMID: 27507154 Free PMC article.
-
CAR1 deletion by CRISPR/Cas9 reduces formation of ethyl carbamate from ethanol fermentation by Saccharomyces cerevisiae.J Ind Microbiol Biotechnol. 2016 Nov;43(11):1517-1525. doi: 10.1007/s10295-016-1831-x. Epub 2016 Aug 29. J Ind Microbiol Biotechnol. 2016. PMID: 27573438
-
[Improvement of inhibitors tolerance of Saccharomyces cerevisiae by overexpressing of long chain sphingoid kinases encoding gene LCB4].Sheng Wu Gong Cheng Xue Bao. 2018 Jun 25;34(6):906-915. doi: 10.13345/j.cjb.170476. Sheng Wu Gong Cheng Xue Bao. 2018. PMID: 29943536 Chinese.
-
Omics analysis of acetic acid tolerance in Saccharomyces cerevisiae.World J Microbiol Biotechnol. 2017 May;33(5):94. doi: 10.1007/s11274-017-2259-9. Epub 2017 Apr 12. World J Microbiol Biotechnol. 2017. PMID: 28405910 Review.
-
[Advances in functional genomics studies underlying acetic acid tolerance of Saccharomyces cerevisiae].Sheng Wu Gong Cheng Xue Bao. 2014 Mar;30(3):368-80. Sheng Wu Gong Cheng Xue Bao. 2014. PMID: 25007573 Review. Chinese.
References
-
- Antar M., Lyu D.M., Nazari M., Shah A.T., Zhou X.M., Smith D.L. Biomass for a sustainable bioeconomy: an overview of world biomass production and utilization. Renew Sustain Energy Rev. 2021;139(18) doi: 10.1016/j.rser.2020.110691. - DOI
-
- Zhao X.Q., Xiong L., Zhang M.M., Bai F.W. Towards efficient bioethanol production from agricultural and forestry residues: exploration of unique natural microorganisms in combination with advanced strain engineering. Bioresour Technol. 2016;215:84–91. doi: 10.1016/j.biortech.2016.03.158. - DOI - PubMed
LinkOut - more resources
Full Text Sources

