IDeRare: a lightweight and extensible open-source phenotype and exome analysis pipeline for germline rare disease diagnosis
- PMID: 38883202
- PMCID: PMC11179852
- DOI: 10.1093/jamiaopen/ooae052
IDeRare: a lightweight and extensible open-source phenotype and exome analysis pipeline for germline rare disease diagnosis
Abstract
Objectives: Diagnosing rare diseases is an arduous and challenging process in clinical settings, resulting in the late discovery of novel variants and referral loops. To help clinicians, we built IDeRare pipelines to accelerate phenotype-genotype analysis for patients with suspected rare diseases.
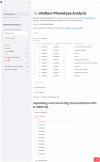
Materials and methods: IDeRare pipeline is separated into phenotype and genotype parts. The phenotype utilizes our handmade Python library, while the genotype part utilizes command line (bash) and Python script to combine bioinformatics executable and Docker image.
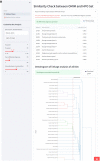
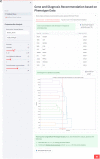
Results: We described various implementations of IDeRare phenotype and genotype parts with real-world clinical and exome data using IDeRare, accelerating the terminology conversion process and giving insight on the diagnostic pathway based on disease linkage analysis until exome analysis and HTML-based reporting for clinicians.
Conclusion: IDeRare is freely available under the BSD-3 license, obtainable via GitHub. The portability of IDeRare pipeline could be easily implemented for semi-technical users and extensible for advanced users.
Keywords: genotype; open-source software; phenotype; rare disease; terminology.
© The Author(s) 2024. Published by Oxford University Press on behalf of the American Medical Informatics Association.
Conflict of interest statement
None declared.
Figures
References
-
- Wright CF, FitzPatrick DR, Firth HV.. Paediatric genomics: diagnosing rare disease in children. Nat Rev Genet. 2018;19(5):253-268. - PubMed
-
- Ministry of Health of Republic of Indonesia. 2024. SATUSEHAT Platform. Accessed May 01, 2024. https://satusehat.kemkes.go.id/platform/docs/id/playbook/
LinkOut - more resources
Full Text Sources