Organic phosphate but not inorganic phosphate regulates Fgf23 expression through MAPK and TGF-ꞵ signaling
- PMID: 38883842
- PMCID: PMC11178987
- DOI: 10.1016/j.isci.2024.109625
Organic phosphate but not inorganic phosphate regulates Fgf23 expression through MAPK and TGF-ꞵ signaling
Abstract
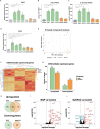
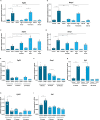
One of the main regulators of phosphate homeostasis is fibroblast growth factor 23 (FGF23), secreted by osteocytes. The effects of organic versus inorganic dietary phosphate on this homeostasis are unclear. This study used MC3T3-E1 FGF23-producing cells to examine the transcriptomic responses to these phosphates. Most importantly, the expression and secretion of FGF23 were only increased in response to organic phosphate. Gene ontology terms related to a response to environmental change were only enriched in cells treated with organic phosphate while cells treated with inorganic phosphate were enriched for terms associated with regulation of cellular phosphate metabolism. Inhibition of MAPK signaling diminished the response of Fgf23 to organic phosphate, suggesting it activates FGF23. TGF-β signaling inhibition increased Fgf23 expression after the addition of organic phosphate, while the negative TGF-β regulator Skil decreased this response. In summary, the observed differential response of FGF23-producing to phosphate types may have consequences for phosphate homeostasis.
Keywords: Transcriptomics; cell biology; molecular biology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

