The Surprising Diversity of UV-Induced Mutations
- PMID: 38884048
- PMCID: PMC11170076
- DOI: 10.1002/ggn2.202300205
The Surprising Diversity of UV-Induced Mutations
Abstract
Ultraviolet (UV) light is the most pervasive environmental mutagen and the primary cause of skin cancer. Genome sequencing of melanomas and other skin cancers has revealed that the vast majority of somatic mutations in these tumors are cytosine-to-thymine (C>T) substitutions in dipyrimidine sequences, which, together with tandem CC>TT substitutions, comprise the canonical UV mutation "signature". These mutation classes are caused by DNA damage directly induced by UV absorption, namely cyclobutane pyrimidine dimers (CPDs) or 6-4 pyrimidine-pyrimidone photoproducts (6-4PP), which form between neighboring pyrimidine bases. However, many of the key driver mutations in melanoma do not fit this mutation signature, but instead are caused by T>A, T>C, C>A, or AC>TT substitutions, frequently occurring in non-dipyrimidine sequence contexts. This article describes recent studies indicating that UV light causes a more diverse spectrum of mutations than previously appreciated, including many of the mutation classes observed in melanoma driver mutations. Potential mechanisms for these diverse mutation signatures are discussed, including UV-induced pyrimidine-purine photoproducts and indirect DNA damage induced by UVA light. Finally, the article reviews recent findings indicating that human DNA polymerase eta normally suppresses these non-canonical UV mutation classes, which can potentially explain why canonical C>T substitutions predominate in human skin cancers.
Keywords: DNA repair; UVA; atypical photoproducts; reactive oxygen species; skin cancer; ultraviolet light.
© 2024 The Authors. Advanced Genetics published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
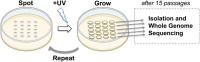
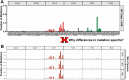

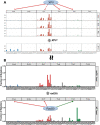

Similar articles
-
Damage mapping techniques and the light they have shed on canonical and atypical UV photoproducts.Front Genet. 2023 Jan 10;13:1102593. doi: 10.3389/fgene.2022.1102593. eCollection 2022. Front Genet. 2023. PMID: 36704334 Free PMC article. Review.
-
Atypical UV Photoproducts Induce Non-canonical Mutation Classes Associated with Driver Mutations in Melanoma.Cell Rep. 2020 Nov 17;33(7):108401. doi: 10.1016/j.celrep.2020.108401. Cell Rep. 2020. PMID: 33207206 Free PMC article.
-
Mechanistic considerations on the wavelength-dependent variations of UVR genotoxicity and mutagenesis in skin: the discrimination of UVA-signature from UV-signature mutation.Photochem Photobiol Sci. 2018 Dec 5;17(12):1861-1871. doi: 10.1039/c7pp00360a. Photochem Photobiol Sci. 2018. PMID: 29850669 Review.
-
[Ultraviolet A-induced DNA damage: role in skin cancer].Bull Acad Natl Med. 2014 Feb;198(2):273-95. Bull Acad Natl Med. 2014. PMID: 26263704 French.
-
Genome-wide maps of UVA and UVB mutagenesis in yeast reveal distinct causative lesions and mutational strand asymmetries.Genetics. 2023 Jul 6;224(3):iyad086. doi: 10.1093/genetics/iyad086. Genetics. 2023. PMID: 37170598 Free PMC article.
Cited by
-
Interplay of replication timing, DNA repair, and translesion synthesis in UV mutagenesis in yeast.Nucleus. 2025 Dec;16(1):2476935. doi: 10.1080/19491034.2025.2476935. Epub 2025 Mar 13. Nucleus. 2025. PMID: 40079129 Free PMC article.
-
Advances in Cell and Immune Therapies for Melanoma.Biomedicines. 2025 Jan 3;13(1):98. doi: 10.3390/biomedicines13010098. Biomedicines. 2025. PMID: 39857682 Free PMC article. Review.
-
Genome-wide maps of UV damage repair and mutation suppression by CPD photolyase.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf495. doi: 10.1093/nar/gkaf495. Nucleic Acids Res. 2025. PMID: 40498072 Free PMC article.
-
Smart Fluids As Autophagy-Activating Photoprotectors: In Vitro Analysis of Dead Sea Water and Magnetized Saline Water Against Ultraviolet B (UVB)-Induced Photodamage in Human Keratinocytes.Cureus. 2025 Apr 14;17(4):e82224. doi: 10.7759/cureus.82224. eCollection 2025 Apr. Cureus. 2025. PMID: 40370877 Free PMC article.
-
Comprehensive Next Generation Sequencing Reveals that Purported Primary Squamous Cell Carcinomas of the Parotid Gland are Genetically Heterogeneous.Head Neck Pathol. 2024 Oct 17;18(1):106. doi: 10.1007/s12105-024-01714-6. Head Neck Pathol. 2024. PMID: 39417927
References
-
- Bonilla X., Parmentier L., King B., Bezrukov F., Kaya G., Zoete V., Seplyarskiy V. B., Sharpe H. J., McKee T., Letourneau A., Ribaux P. G., Popadin K., Basset‐Seguin N., Ben Chaabene R., Santoni F. A., Andrianova M. A., Guipponi M., Garieri M., Verdan C., Grosdemange K., Sumara O., Eilers M., Aifantis I., Michielin O., de Sauvage F. J., Antonarakis S. E., Nikolaev S. I., Nat. Genet. 2016, 48, 398. - PubMed
-
- Hodis E., Watson I. R., Kryukov G. V., Arold S. T., Imielinski M., Theurillat J. P., Nickerson E., Auclair D., Li L., Place C., Dicara D., Ramos A. H., Lawrence M. S., Cibulskis K., Sivachenko A., Voet D., Saksena G., Stransky N., Onofrio R. C., Winckler W., Ardlie K., Wagle N., Wargo J., Chong K., Morton D. L., Stemke‐Hale K., Chen G., Noble M., Meyerson M., Ladbury J. E., et al., Cell 2012, 150, 251. - PMC - PubMed
-
- Hayward N. K., Wilmott J. S., Waddell N., Johansson P. A., Field M. A., Nones K., Patch A. M., Kakavand H., Alexandrov L. B., Burke H., Jakrot V., Kazakoff S., Holmes O., Leonard C., Sabarinathan R., Mularoni L., Wood S., Xu Q., Waddell N., Tembe V., Pupo G. M., De Paoli‐Iseppi R., Vilain R. E., Shang P., Lau L. M. S., Dagg R. A., Schramm S. J., Pritchard A., Dutton‐Regester K., Newell F., et al., Nature 2017, 545, 175. - PubMed
-
- Pfeifer G. P., You Y. H., Besaratinia A., Mutat. Res. 2005, 571, 19. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
