Clinical Application of Polygenic Risk Score in IgA Nephropathy
- PMID: 38884057
- PMCID: PMC11169313
- DOI: 10.1007/s43657-023-00138-6
Clinical Application of Polygenic Risk Score in IgA Nephropathy
Abstract
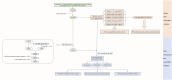
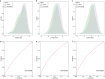
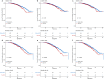
Genome-wide association studies (GWASs) have identified 30 independent genetic variants associated with IgA nephropathy (IgAN). A genetic risk score (GRS) represents the number of risk alleles carried and thus captures an individual's genetic risk. However, whether and which polygenic risk score crucial for the evaluation of any potential personal or clinical utility on risk and prognosis are still obscure. We constructed different GRS models based on different sets of variants, which were top single nucleotide polymorphisms (SNPs) reported in the previous GWASs. The case-control GRS analysis included 3365 IgAN patients and 8842 healthy individuals. The association between GRS and clinical variability, including age at diagnosis, clinical parameters, Oxford pathology classification, and kidney prognosis was further evaluated in a prospective cohort of 1747 patients. Three GRS models (15 SNPs, 21 SNPs, and 55 SNPs) were constructed after quality control. The patients with the top 20% GRS had 2.42-(15 SNPs, p = 8.12 × 10-40), 3.89-(21 SNPs, p = 3.40 × 10-80) and 3.73-(55 SNPs, p = 6.86 × 10-81) fold of risk to develop IgAN compared to the patients with the bottom 20% GRS, with area under the receiver operating characteristic curve (AUC) of 0.59, 0.63, and 0.63 in group discriminations, respectively. A positive correlation between GRS and microhematuria, mesangial hypercellularity, segmental glomerulosclerosis and a negative correlation on the age at diagnosis, body mass index (BMI), mean arterial pressure (MAP), serum C3, triglycerides can be observed. Patients with the top 20% GRS also showed a higher risk of worse prognosis for all three models (1.36, 1.42, and 1.36 fold of risk) compared to the remaining 80%, whereas 21 SNPs model seemed to show a slightly better fit in prediction. Collectively, a higher burden of risk variants is associated with earlier disease onset and a higher risk of a worse prognosis. This may be informational in translating knowledge on IgAN genetics into disease risk prediction and patient stratification.
Supplementary information: The online version contains supplementary material available at 10.1007/s43657-023-00138-6.
Keywords: Genomics; IgA nephropathy; Polygenic score; Prognosis; Risk prediction.
© International Human Phenome Institutes (Shanghai) 2024. Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestOn behalf of all authors, the corresponding author states that there is no conflict of interest.
Figures
Similar articles
-
IgA Nephropathy Genetic Risk Score to Estimate the Prevalence of IgA Nephropathy in UK Biobank.Kidney Int Rep. 2020 Jul 19;5(10):1643-1650. doi: 10.1016/j.ekir.2020.07.012. eCollection 2020 Oct. Kidney Int Rep. 2020. PMID: 33102956 Free PMC article.
-
Assessing the clinical utility of a genetic risk score constructed using 49 susceptibility alleles for type 2 diabetes in a Japanese population.J Clin Endocrinol Metab. 2013 Oct;98(10):E1667-73. doi: 10.1210/jc.2013-1642. Epub 2013 Aug 16. J Clin Endocrinol Metab. 2013. PMID: 23956346
-
Body mass index interacts with a genetic-risk score for depression increasing the risk of the disease in high-susceptibility individuals.Transl Psychiatry. 2022 Jan 24;12(1):30. doi: 10.1038/s41398-022-01783-7. Transl Psychiatry. 2022. PMID: 35075110 Free PMC article.
-
Oxford Classification of IgA nephropathy 2016: an update from the IgA Nephropathy Classification Working Group.Kidney Int. 2017 May;91(5):1014-1021. doi: 10.1016/j.kint.2017.02.003. Epub 2017 Mar 22. Kidney Int. 2017. PMID: 28341274 Review.
-
Polygenic risk prediction models for colorectal cancer: a systematic review.BMC Cancer. 2022 Jan 15;22(1):65. doi: 10.1186/s12885-021-09143-2. BMC Cancer. 2022. PMID: 35030997 Free PMC article.
Cited by
-
Towards Risk Stratification in Clinical Care for IgA Nephropathy: Genetic Risk Scores: Comments on the Study "Clinical Application of Polygenic Risk Score in IgA Nephropathy".Phenomics. 2024 Nov 12;4(5):527-530. doi: 10.1007/s43657-024-00184-8. eCollection 2024 Oct. Phenomics. 2024. PMID: 39723228 No abstract available.
-
Navigating Genetic Testing in Nephrology: Options and Decision-Making Strategies.Kidney Int Rep. 2024 Dec 27;10(3):673-695. doi: 10.1016/j.ekir.2024.12.020. eCollection 2025 Mar. Kidney Int Rep. 2024. PMID: 40225372 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
