The astrocyte-enriched gene deathstar plays a crucial role in the development, locomotion, and lifespan of D. melanogaster
- PMID: 38884422
- PMCID: PMC11185185
- DOI: 10.1080/19336934.2024.2368336
The astrocyte-enriched gene deathstar plays a crucial role in the development, locomotion, and lifespan of D. melanogaster
Abstract
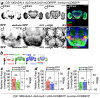
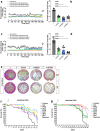
The Drosophila melanogaster brain is a complex organ with various cell types, orchestrating the development, physiology, and behaviors of the fly. While each cell type in Drosophila brain is known to express a unique gene set, their complete genetic profile is still unknown. Advances in the RNA sequencing techniques at single-cell resolution facilitate identifying novel cell type markers and/or re-examining the specificity of the available ones. In this study, exploiting a single-cell RNA sequencing data of Drosophila optic lobe, we categorized the cells based on their expression pattern for known markers, then the genes with enriched expression in astrocytes were identified. CG11000 was identified as a gene with a comparable expression profile to the Eaat1 gene, an astrocyte marker, in every individual cell inside the Drosophila optic lobe and midbrain, as well as in the entire Drosophila brain throughout its development. Consistent with our bioinformatics data, immunostaining of the brains dissected from transgenic adult flies showed co-expression of CG11000 with Eaat1 in a set of single cells corresponding to the astrocytes in the Drosophila brain. Physiologically, inhibiting CG11000 through RNA interference disrupted the normal development of male D. melanogaster, while having no impact on females. Expression suppression of CG11000 in adult flies led to decreased locomotion activity and also shortened lifespan specifically in astrocytes, indicating the gene's significance in astrocytes. We designated this gene as 'deathstar' due to its crucial role in maintaining the star-like shape of glial cells, astrocytes, throughout their development into adult stage.
Keywords: CG11000; Eaat1; astrocyte; deathstar; glia; male development; single-cell RNA sequencing.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
