Oncogenic Calreticulin Induces Immune Escape by Stimulating TGFβ Expression and Regulatory T-cell Expansion in the Bone Marrow Microenvironment
- PMID: 38885318
- PMCID: PMC11405138
- DOI: 10.1158/0008-5472.CAN-23-3553
Oncogenic Calreticulin Induces Immune Escape by Stimulating TGFβ Expression and Regulatory T-cell Expansion in the Bone Marrow Microenvironment
Abstract
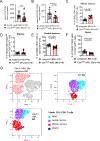
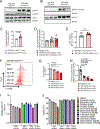
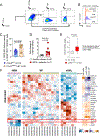
Increasing evidence supports the interplay between oncogenic mutations and immune escape mechanisms. Strategies to counteract the immune escape mediated by oncogenic signaling could provide improved therapeutic options for patients with various malignancies. As mutant calreticulin (CALR) is a common driver of myeloproliferative neoplasms (MPN), we analyzed the impact of oncogenic CALRdel52 on the bone marrow (BM) microenvironment in MPN. Single-cell RNA sequencing revealed that CALRdel52 led to the expansion of TGFβ1-producing erythroid progenitor cells and promoted the expansion of FoxP3+ regulatory T cells (Treg) in a murine MPN model. Treatment with an anti-TGFβ antibody improved mouse survival and increased the glycolytic activity in CD4+ and CD8+ T cells in vivo, whereas T-cell depletion abrogated the protective effects conferred by neutralizing TGFβ. TGFβ1 reduced perforin and TNFα production by T cells in vitro. TGFβ1 production by CALRdel52 cells was dependent on JAK1/2, PI3K, and ERK activity, which activated the transcription factor Sp1 to induce TGFβ1 expression. In four independent patient cohorts, TGFβ1 expression was increased in the BM of patients with MPN compared with healthy individuals, and the BM of patients with MPN contained a higher frequency of Treg compared with healthy individuals. Together, this study identified an ERK/Sp1/TGFβ1 axis in CALRdel52 MPNs as a mechanism of immunosuppression that can be targeted to elicit T-cell-mediated cytotoxicity. Significance: Targeting the mutant calreticulin/TGFβ1 axis increases T-cell activity and glycolytic capacity, providing the rationale for conducting clinical trials on TGFβ antagonists as an immunotherapeutic strategy in patients with myeloproliferative neoplasms.
©2024 American Association for Cancer Research.
Conflict of interest statement
Figures





References
-
- Prestipino A, Zeiser R. Clinical implications of tumor-intrinsic mechanisms regulating PD-L1. Science translational medicine 2019;11(478):eaav4810. - PubMed
-
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. New England Journal of Medicine 2013;369(25):2379–90. - PubMed
MeSH terms
Substances
Grants and funding
- R01 HL095791/HL/NHLBI NIH HHS/United States
- P01 CA065493/CA/NCI NIH HHS/United States
- R01 HL118979/HL/NHLBI NIH HHS/United States
- CIBSS -/Deutsche Forschungsgemeinschaft (DFG)
- KL2 TR000113/TR/NCATS NIH HHS/United States
- R37 AI034495/AI/NIAID NIH HHS/United States
- ERC-2022-ADG Project: 101094168/HORIZON EUROPE European Research Council (ERC)
- DJCLS 09R/2022/Jose-Carreras Leukemia Foundation
- 101094168/ERC_/European Research Council/International
- P01 HL158505/HL/NHLBI NIH HHS/United States
- 70113473/Deutsche Krebshilfe (German Cancer Aid)
- DFG Research Unit RU5659 (TARGET MPN)/Deutsche Forschungsgemeinschaft (DFG)
- SFB-1479 -/Deutsche Forschungsgemeinschaft (DFG)
- PM4Onco FKZ 01ZZ2322A/German Federal Ministry of Education and Research
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

