Iron-(Fe3+)-Dependent Reactivation of Telomerase Drives Colorectal Cancers
- PMID: 38885349
- PMCID: PMC11450372
- DOI: 10.1158/2159-8290.CD-23-1379
Iron-(Fe3+)-Dependent Reactivation of Telomerase Drives Colorectal Cancers
Abstract
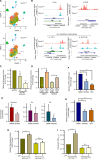
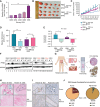
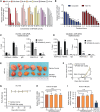
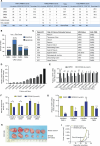
Over-consumption of iron-rich red meat and hereditary or genetic iron overload are associated with an increased risk of colorectal carcinogenesis, yet the mechanistic basis of how metal-mediated signaling leads to oncogenesis remains enigmatic. Using fresh colorectal cancer samples we identify Pirin, an iron sensor, that overcomes a rate-limiting step in oncogenesis, by reactivating the dormant human telomerase reverse transcriptase (hTERT) subunit of the telomerase holoenzyme in an iron-(Fe3+)-dependent manner and thereby drives colorectal cancers. Chemical genetic screens combined with isothermal dose-response fingerprinting and mass spectrometry identified a small molecule SP2509 that specifically inhibits Pirin-mediated hTERT reactivation in colorectal cancers by competing with iron-(Fe3+) binding. Our findings, first to document how metal ions reactivate telomerase, provide a molecular mechanism for the well-known association between red meat and increased incidence of colorectal cancers. Small molecules like SP2509 represent a novel modality to target telomerase that acts as a driver of 90% of human cancers and is yet to be targeted in clinic. Significance: We show how iron-(Fe3+) in collusion with genetic factors reactivates telomerase, providing a molecular mechanism for the association between iron overload and increased incidence of colorectal cancers. Although no enzymatic inhibitors of telomerase have entered the clinic, we identify SP2509, a small molecule that targets telomerase reactivation and function in colorectal cancers.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
R. Shanmugam reports a patent for Specific inhibition of telomerase activity in colorectal cancer pending. J.-L. Low reports grants from National Research Foundation, Singapore, during the conduct of the study. I.B. Tan reports grants and personal fees from MSD and Roche, personal fees from BMS, AstraZeneca, and Merck Serano, and personal fees from Amgen outside the submitted work; in addition, I.B. Tan has a patent for related to current work pending. V. Tergaonkar reports grants from National Research Foundation (NRF) and National Medical Research Council (NMRC) during the conduct of the study. No disclosures were reported by the other authors.
Figures


References
-
- Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019;16:713–32. - PubMed
-
- Sato Y, Nakaya N, Kuriyama S, Nishino Y, Tsubono Y, Tsuji I. Meat consumption and risk of colorectal cancer in Japan: the Miyagi Cohort Study. Eur J Cancer Prev 2006;15:211–8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

