NCF4 attenuates colorectal cancer progression by modulating inflammasome activation and immune surveillance
- PMID: 38886341
- PMCID: PMC11183137
- DOI: 10.1038/s41467-024-49549-7
NCF4 attenuates colorectal cancer progression by modulating inflammasome activation and immune surveillance
Abstract
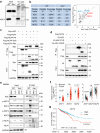
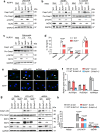
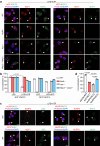
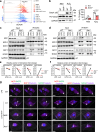
The spatiotemporal regulation of inflammasome activation remains unclear. To examine the mechanism underlying the assembly and regulation of the inflammasome response, here we perform an immunoprecipitation-mass spectrometry analysis of apoptosis-associated speck-like protein containing a CARD (ASC) and identify NCF4/1/2 as ASC-binding proteins. Reduced NCF4 expression is associated with colorectal cancer development and decreased five-year survival rate in patients with colorectal cancer. NCF4 cooperates with NCF1 and NCF2 to promote NLRP3 and AIM2 inflammasome activation. Mechanistically, NCF4 phosphorylation and puncta distribution switches from the NADPH complex to the perinuclear region, mediating ASC oligomerization, speck formation and inflammasome activation. NCF4 functions as a sensor of ROS levels, to establish a balance between ROS production and inflammasome activation. NCF4 deficiency causes severe colorectal cancer in mice, increases transit-amplifying and precancerous cells, reduces the frequency and activation of CD8+ T and NK cells, and impairs the inflammasome-IL-18-IFN-γ axis during the early phase of colorectal tumorigenesis. Our study implicates NCF4 in determining the spatial positioning of inflammasome assembly and contributing to inflammasome-mediated anti-tumor responses.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

