Remodeling of anti-tumor immunity with antibodies targeting a p53 mutant
- PMID: 38886748
- PMCID: PMC11184848
- DOI: 10.1186/s13045-024-01566-1
Remodeling of anti-tumor immunity with antibodies targeting a p53 mutant
Abstract
Background: p53, the most frequently mutated gene in cancer, lacks effective targeted drugs.
Methods: We developed monoclonal antibodies (mAbs) that target a p53 hotspot mutation E285K without cross-reactivity with wild-type p53. They were delivered using lipid nanoparticles (LNPs) that encapsulate DNA plasmids. Western blot, BLI, flow cytometry, single-cell sequencing (scRNA-seq), and other methods were employed to assess the function of mAbs in vitro and in vivo.
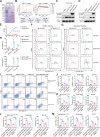
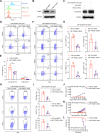
Results: These LNP-pE285K-mAbs in the IgG1 format exhibited a robust anti-tumor effect, facilitating the infiltration of immune cells, including CD8+ T, B, and NK cells. scRNA-seq revealed that IgG1 reduces immune inhibitory signaling, increases MHC signaling from B cells to CD8+ T cells, and enriches anti-tumor T cell and B cell receptor profiles. The E285K-mAbs were also produced in the dimeric IgA (dIgA) format, whose anti-tumor activity depended on the polymeric immunoglobulin receptor (PIGR), a membrane Ig receptor, whereas that of IgG1 relied on TRIM21, an intracellular IgG receptor.
Conclusions: Targeting specific mutant epitopes using DNA-encoded and LNP-delivered mAbs represents a potential precision medicine strategy against p53 mutants in TRIM21- or PIGR-positive cancers.
Keywords: E285K; IgG1; Monoclonal antibody; Mutant p53; dIgA.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

