Approach to the Patient With Suspected Silver-Russell Syndrome
- PMID: 38888172
- PMCID: PMC11403326
- DOI: 10.1210/clinem/dgae423
Approach to the Patient With Suspected Silver-Russell Syndrome
Abstract
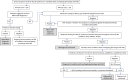
Silver-Russell syndrome (SRS) is a clinical diagnosis requiring the fulfillment of ≥ 4/6 Netchine-Harbison Clinical Scoring System (NH-CSS) criteria. A score of ≥ 4/6 NH-CSS (or ≥ 3/6 with strong clinical suspicion) warrants (epi)genetic confirmation, identifiable in ∼60% patients. The approach to the investigation and diagnosis of SRS is detailed in the only international consensus guidance, published in 2016. In the intervening years, the clinical, biochemical, and (epi)genetic characteristics of SRS have rapidly expanded, largely attributable to advancing molecular genetic techniques and a greater awareness of related disorders. The most common etiologies of SRS remain loss of methylation of chromosome 11p15 (11p15LOM) and maternal uniparental disomy of chromosome 7 (upd(7)mat). Rarer causes of SRS include monogenic pathogenic variants in imprinted (CDKN1C and IGF2) and non-imprinted (PLAG1 and HMGA2) genes. Although the age-specific NH-CSS can identify more common molecular causes of SRS, its use in identifying monogenic causes is unclear. Preliminary data suggest that NH-CSS is poor at identifying many of these cases. Additionally, there has been increased recognition of conditions with phenotypes overlapping with SRS that may fulfill NH-CSS criteria but have distinct genetic etiologies and disease trajectories. This group of conditions is frequently overlooked and under-investigated, leading to no or delayed diagnosis. Like SRS, these conditions are multisystemic disorders requiring multidisciplinary care and tailored management strategies. Early identification is crucial to improve outcomes and reduce the major burden of the diagnostic odyssey for patients and families. This article aims to enable clinicians to identify key features of rarer causes of SRS and conditions with overlapping phenotypes, show a logical approach to the molecular investigation, and highlight the differences in clinical management strategies.
Keywords: NH-CSS; Silver-Russell syndrome; diagnosis; genetic.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Endocrine Society.
Figures

References
-
- Walenkamp MJE, Robers JML, Wit JM, et al. . Phenotypic features and response to GH treatment of patients with a molecular defect of the IGF-1 receptor. J Clin Endocrinol Metab. 2019;104(8):3157‐3171. - PubMed
-
- Wakeling EL, Brioude F, Lokulo-Sodipe O, et al. . Diagnosis and management of Silver–Russell syndrome: first international consensus statement. Nat Rev Endocrinol. 2017;13(2):105‐124. - PubMed
-
- Bakker B, Sonneveld LJH, Woltering MC, Bikker H, Kant SG. A girl with Beckwith-Wiedemann syndrome and pseudohypoparathyroidism type 1B due to multiple imprinting defects. J Clin Endocrinol Metab. 2015;100(11):3963‐3966. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

