The crosstalk role of CDKN2A between tumor progression and cuproptosis resistance in colorectal cancer
- PMID: 38888512
- PMCID: PMC11236303
- DOI: 10.18632/aging.205945
The crosstalk role of CDKN2A between tumor progression and cuproptosis resistance in colorectal cancer
Abstract
Background: Cuproptosis is a type of cell death characterized by excessive copper-lipid reactions in the tricarboxylic acid cycle, resulting in protein toxicity stress and cell death. Although known as a cuproptosis inhibitor through CRISPR-Cas9 screening, the role of cyclin-dependent kinase inhibitor 2A (CDKN2A) in cuproptosis resistance and its connection to tumor development remains unclear.
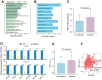
Methods: In this study, we combined single-cell sequencing, spatial transcriptomics, pathological image analysis, TCGA multi-omics analysis and in vitro experimental validation to comprehensively investigate CDKN2A distribution, expression, epigenetic modification, regulation and genomic features in colorectal cancer cells. We further explored the associations between CDKN2A and cellular pathway, immune infiltration and spatial signal communication.
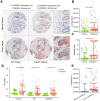
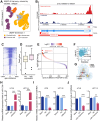
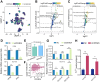
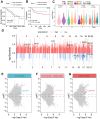
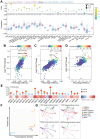
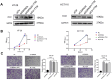
Results: Our findings showed an increasing trend in cuproptosis in the trajectory of tumor progression, accompanied by an upward trend of CDKN2A. CDKN2A underwent transcriptional activation by MEF2D and via the SNHG7/miR-133b axis, upregulating glycolysis, copper metabolism and copper ion efflux. CDKN2A likely drives epithelial-mesenchymal transition (EMT) and progression by activating Wnt signaling. CDKN2A is associated with high genomic instability and sensitivity to radiation and chemotherapy. Tumor regions expressing CDKN2A exhibit distinctive SPP1+ tumor-associated macrophage (TAM) infiltration and MMP7 enrichment, along with unique signaling crosstalk with adjacent areas.
Conclusions: CDKN2A mediates cuproptosis resistance through regulating glycolysis and copper homeostasis, accompanied by a malignant phenotype and pro-tumor niche. Radiation and chemotherapy are expected to potentially serve as therapeutic approaches for cuproptosis-resistant colorectal cancer with high CDKN2A expression.
Keywords: CDKN2A; bioinformatics; cuproptosis; metabolism; radiation therapy.
Conflict of interest statement
Figures


Similar articles
-
Comprehensive analyses of cuproptosis-related gene CDKN2A on prognosis and immunologic therapy in human tumors.Medicine (Baltimore). 2023 Apr 7;102(14):e33468. doi: 10.1097/MD.0000000000033468. Medicine (Baltimore). 2023. PMID: 37026918 Free PMC article.
-
A Cuproptosis-Related gene Signature as a Prognostic Biomarker in Thyroid Cancer Based on Transcriptomics.Biochem Genet. 2025 Apr;63(2):1584-1604. doi: 10.1007/s10528-024-10767-9. Epub 2024 Apr 9. Biochem Genet. 2025. PMID: 38594571
-
Cuproptosis-related gene ACAD8 inhibits the metastatic ability of colorectal cancer by inducing cuproptosis.Front Immunol. 2025 Apr 3;16:1560322. doi: 10.3389/fimmu.2025.1560322. eCollection 2025. Front Immunol. 2025. PMID: 40248707 Free PMC article.
-
Cuproptosis regulation by long noncoding RNAs: Mechanistic insights and clinical implications in cancer.Arch Biochem Biophys. 2025 Mar;765:110324. doi: 10.1016/j.abb.2025.110324. Epub 2025 Feb 1. Arch Biochem Biophys. 2025. PMID: 39900259 Review.
-
Cuproptosis: A novel therapeutic target for overcoming cancer drug resistance.Drug Resist Updat. 2024 Jan;72:101018. doi: 10.1016/j.drup.2023.101018. Epub 2023 Nov 11. Drug Resist Updat. 2024. PMID: 37979442 Review.
Cited by
-
Cancer therapy resistance from a spatial-omics perspective.Clin Transl Med. 2025 Jul;15(7):e70396. doi: 10.1002/ctm2.70396. Clin Transl Med. 2025. PMID: 40677104 Free PMC article. Review.
-
Copper homeostasis and copper-induced cell death in tumor immunity: implications for therapeutic strategies in cancer immunotherapy.Biomark Res. 2024 Oct 31;12(1):130. doi: 10.1186/s40364-024-00677-8. Biomark Res. 2024. PMID: 39482784 Free PMC article. Review.
-
Cuproptosis-related genes and agents: implications in tumor drug resistance and future perspectives.Front Pharmacol. 2025 May 8;16:1559236. doi: 10.3389/fphar.2025.1559236. eCollection 2025. Front Pharmacol. 2025. PMID: 40406488 Free PMC article. Review.
-
Big data analysis and machine learning of the role of cuproptosis-related long non-coding RNAs (CuLncs) in the prognosis and immune landscape of ovarian cancer.Front Immunol. 2025 Feb 25;16:1555782. doi: 10.3389/fimmu.2025.1555782. eCollection 2025. Front Immunol. 2025. PMID: 40070821 Free PMC article.
-
The important role of cuproptosis and cuproptosis-related genes in the development of thyroid carcinoma revealed by transcriptomic analysis and experiments.Braz J Otorhinolaryngol. 2025 May-Jun;91(3):101560. doi: 10.1016/j.bjorl.2025.101560. Epub 2025 Feb 5. Braz J Otorhinolaryngol. 2025. PMID: 39914044 Free PMC article.
References
-
- Provenzale D, Ness RM, Llor X, Weiss JM, Abbadessa B, Cooper G, Early DS, Friedman M, Giardiello FM, Glaser K, Gurudu S, Halverson AL, Issaka R, et al.. NCCN Guidelines Insights: Colorectal Cancer Screening, Version 2.2020. J Natl Compr Canc Netw. 2020; 18:1312–20. 10.6004/jnccn.2020.0048 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

