Elucidating the Role of Human ALAS2 C-terminal Mutations Resulting in Loss of Function and Disease
- PMID: 38888931
- PMCID: PMC11223264
- DOI: 10.1021/acs.biochem.4c00066
Elucidating the Role of Human ALAS2 C-terminal Mutations Resulting in Loss of Function and Disease
Abstract
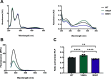
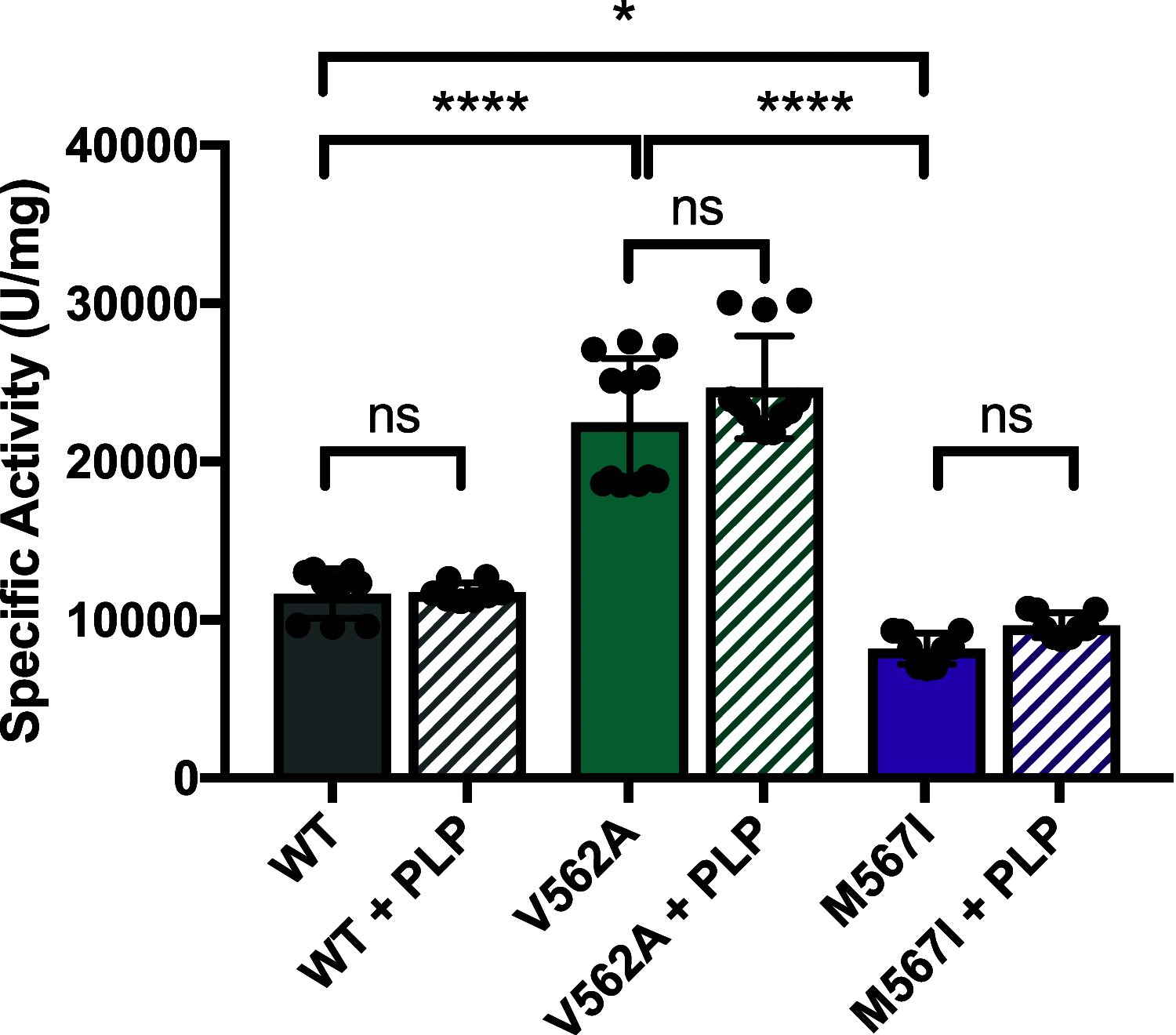
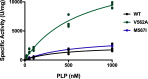
The conserved enzyme aminolevulinic acid synthase (ALAS) initiates heme biosynthesis in certain bacteria and eukaryotes by catalyzing the condensation of glycine and succinyl-CoA to yield aminolevulinic acid. In humans, the ALAS isoform responsible for heme production during red blood cell development is the erythroid-specific ALAS2 isoform. Owing to its essential role in erythropoiesis, changes in human ALAS2 (hALAS2) function can lead to two different blood disorders. X-linked sideroblastic anemia results from loss of ALAS2 function, while X-linked protoporphyria results from gain of ALAS2 function. Interestingly, mutations in the ALAS2 C-terminal extension can be implicated in both diseases. Here, we investigate the molecular basis for enzyme dysfunction mediated by two previously reported C-terminal loss-of-function variants, hALAS2 V562A and M567I. We show that the mutations do not result in gross structural perturbations, but the enzyme stability for V562A is decreased. Additionally, we show that enzyme stability moderately increases with the addition of the pyridoxal 5'-phosphate (PLP) cofactor for both variants. The variants display differential binding to PLP and the individual substrates compared to wild-type hALAS2. Although hALAS2 V562A is a more active enzyme in vitro, it is less efficient concerning succinyl-CoA binding. In contrast, the M567I mutation significantly alters the cooperativity of substrate binding. In combination with previously reported cell-based studies, our work reveals the molecular basis by which hALAS2 C-terminal mutations negatively affect ALA production necessary for proper heme biosynthesis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

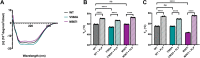
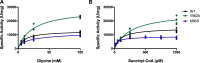
Similar articles
-
Anti-Correlation between the Dynamics of the Active Site Loop and C-Terminal Tail in Relation to the Homodimer Asymmetry of the Mouse Erythroid 5-Aminolevulinate Synthase.Int J Mol Sci. 2018 Jun 28;19(7):1899. doi: 10.3390/ijms19071899. Int J Mol Sci. 2018. PMID: 29958424 Free PMC article.
-
Isoniazid inhibits human erythroid 5-aminolevulinate synthase: Molecular mechanism and tolerance study with four X-linked protoporphyria patients.Biochim Biophys Acta Mol Basis Dis. 2017 Feb;1863(2):428-439. doi: 10.1016/j.bbadis.2016.11.011. Epub 2016 Nov 10. Biochim Biophys Acta Mol Basis Dis. 2017. PMID: 27838491
-
Human Erythroid 5-Aminolevulinate Synthase Mutations Associated with X-Linked Protoporphyria Disrupt the Conformational Equilibrium and Enhance Product Release.Biochemistry. 2015 Sep 15;54(36):5617-31. doi: 10.1021/acs.biochem.5b00407. Epub 2015 Sep 2. Biochemistry. 2015. PMID: 26300302 Free PMC article.
-
Regulation and tissue-specific expression of δ-aminolevulinic acid synthases in non-syndromic sideroblastic anemias and porphyrias.Mol Genet Metab. 2019 Nov;128(3):190-197. doi: 10.1016/j.ymgme.2019.01.015. Epub 2019 Jan 23. Mol Genet Metab. 2019. PMID: 30737140 Review.
-
Structural basis for dysregulation of aminolevulinic acid synthase in human disease.J Biol Chem. 2022 Mar;298(3):101643. doi: 10.1016/j.jbc.2022.101643. Epub 2022 Jan 28. J Biol Chem. 2022. PMID: 35093382 Free PMC article. Review.
References
-
- Greer J. P.; Arber D. A.; List A. F.; Foerster J.. Wintrobe’s Clinical Hematology, 13th ed.; Wolters Kluwer, Lippincott Williams & Wilkins Health: Philadelphia, 2014.
-
- Ducamp S.; Kannengiesser C.; Touati M.; Garcon L.; Guerci-Bresler A.; Guichard J. F.; Vermylen C.; Dochir J.; Poirel H. A.; Fouyssac F.; Mansuy L.; Leroux G.; Tertian G.; Girot R.; Heimpel H.; Matthes T.; Talbi N.; Deybach J. C.; Beaumont C.; Puy H.; Grandchamp B. Sideroblastic anemia: molecular analysis of the ALAS2 gene in a series of 29 probands and functional studies of 10 missense mutations. Hum. Mutat. 2011, 32, 590–597. 10.1002/humu.21455. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

