Short-term cold exposure induces persistent epigenomic memory in brown fat
- PMID: 38889724
- PMCID: PMC11305953
- DOI: 10.1016/j.cmet.2024.05.011
Short-term cold exposure induces persistent epigenomic memory in brown fat
Abstract
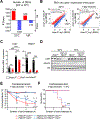
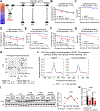
Deficiency of the epigenome modulator histone deacetylase 3 (HDAC3) in brown adipose tissue (BAT) impairs the ability of mice to survive in near-freezing temperatures. Here, we report that short-term exposure to mild cold temperature (STEMCT: 15°C for 24 h) averted lethal hypothermia of mice lacking HDAC3 in BAT (HDAC3 BAT KO) exposed to 4°C. STEMCT restored the induction of the thermogenic coactivator PGC-1α along with UCP1 at 22°C, which is greatly impaired in HDAC3-deficient BAT, and deletion of either UCP1 or PGC-1α prevented the protective effect of STEMCT. Remarkably, this protection lasted for up to 7 days. Transcriptional activator C/EBPβ was induced by short-term cold exposure in mouse and human BAT and, uniquely, remained high for 7 days following STEMCT. Adeno-associated virus-mediated knockdown of BAT C/EBPβ in HDAC3 BAT KO mice erased the persistent memory of STEMCT, revealing the existence of a C/EBPβ-dependent and HDAC3-independent cold-adaptive epigenomic memory.
Keywords: C/EBPβ; ERRα; HDAC3; PGC-1α; UCP1; brown adipose tissue; cold memory; mitochondria; oxidative phosphorylation; thermogenesis.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.A.L. is an advisory board member for Pfizer Inc. and is an advisory board member and co-founder of Flare Therapeutics. S.K. serves on Scientific Advisory Boards for Merck, Alnylum, AbbVie, and Boehringer Ingelheim.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

