H19 influenza A virus exhibits species-specific MHC class II receptor usage
- PMID: 38889725
- PMCID: PMC11295516
- DOI: 10.1016/j.chom.2024.05.018
H19 influenza A virus exhibits species-specific MHC class II receptor usage
Abstract
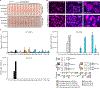
Avian influenza A virus (IAV) surveillance in Northern California, USA, revealed unique IAV hemagglutinin (HA) genome sequences in cloacal swabs from lesser scaups. We found two closely related HA sequences in the same duck species in 2010 and 2013. Phylogenetic analyses suggest that both sequences belong to the recently discovered H19 subtype, which thus far has remained uncharacterized. We demonstrate that H19 does not bind the canonical IAV receptor sialic acid (Sia). Instead, H19 binds to the major histocompatibility complex class II (MHC class II), which facilitates viral entry. Unlike the broad MHC class II specificity of H17 and H18 from bat IAV, H19 exhibits a species-specific MHC class II usage that suggests a limited host range and zoonotic potential. Using cell lines overexpressing MHC class II, we rescued recombinant H19 IAV. We solved the H19 crystal structure and identified residues within the putative Sia receptor binding site (RBS) that impede Sia-dependent entry.
Keywords: HA structure; MHC class II; avian influenza virus surveillance; entry receptor; glycan array; hemagglutinin subtype H19; host range; influenza A virus; receptor binding site; recombinant H19 influenza A virus.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The A.G.-S. laboratory has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories, and Merck outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus, Pfizer, and Prosetta, outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott, and Astrazeneca. A.G.-S. is an inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays, NDV-based SARS-CoV-2 vaccines, influenza virus vaccines, and influenza virus therapeutics, which list F.K. as co-inventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2 and another company, Castlevax, to develop SARS-CoV-2 vaccines. F.K. is a co-founder and scientific advisory board member of Castlevax. F.K. has consulted for Merck, Curevac, Seqirus, and Pfizer and is currently consulting for 3rd Rock Ventures, GSK, Gritstone, and Avimex. The Krammer laboratory is also collaborating with Dynavax on influenza vaccine development.
Figures
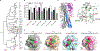


References
-
- Long JS, Mistry B, Haslam SM, and Barclay WS (2019). Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17, 67–81. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

