Digital telomere measurement by long-read sequencing distinguishes healthy aging from disease
- PMID: 38890274
- PMCID: PMC11189511
- DOI: 10.1038/s41467-024-49007-4
Digital telomere measurement by long-read sequencing distinguishes healthy aging from disease
Abstract
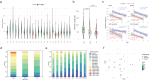
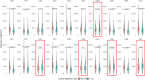
Telomere length is an important biomarker of organismal aging and cellular replicative potential, but existing measurement methods are limited in resolution and accuracy. Here, we deploy digital telomere measurement (DTM) by nanopore sequencing to understand how distributions of human telomere length change with age and disease. We measure telomere attrition and de novo elongation with up to 30 bp resolution in genetically defined populations of human cells, in blood cells from healthy donors and in blood cells from patients with genetic defects in telomere maintenance. We find that human aging is accompanied by a progressive loss of long telomeres and an accumulation of shorter telomeres. In patients with defects in telomere maintenance, the accumulation of short telomeres is more pronounced and correlates with phenotypic severity. We apply machine learning to train a binary classification model that distinguishes healthy individuals from those with telomere biology disorders. This sequencing and bioinformatic pipeline will advance our understanding of telomere maintenance mechanisms and the use of telomere length as a clinical biomarker of aging and disease.
© 2024. The Author(s).
Conflict of interest statement
S. Sanchez and S.E.A. are listed as inventors on a provisional patent application related to clinical applications of telomere measurement related to this work (STFD-006-P). The remaining authors declare no competing interests.
Figures


Update of
-
Digital telomere measurement by long-read sequencing distinguishes healthy aging from disease.bioRxiv [Preprint]. 2023 Dec 1:2023.11.29.569263. doi: 10.1101/2023.11.29.569263. bioRxiv. 2023. Update in: Nat Commun. 2024 Jun 18;15(1):5148. doi: 10.1038/s41467-024-49007-4. PMID: 38077053 Free PMC article. Updated. Preprint.
Similar articles
-
Digital telomere measurement by long-read sequencing distinguishes healthy aging from disease.bioRxiv [Preprint]. 2023 Dec 1:2023.11.29.569263. doi: 10.1101/2023.11.29.569263. bioRxiv. 2023. Update in: Nat Commun. 2024 Jun 18;15(1):5148. doi: 10.1038/s41467-024-49007-4. PMID: 38077053 Free PMC article. Updated. Preprint.
-
Telomeres are elongated in older individuals in a hibernating rodent, the edible dormouse (Glis glis).Sci Rep. 2016 Nov 24;6:36856. doi: 10.1038/srep36856. Sci Rep. 2016. PMID: 27883035 Free PMC article.
-
Telomere shortening and accelerated aging in COPD: findings from the BODE cohort.Respir Res. 2017 Apr 13;18(1):59. doi: 10.1186/s12931-017-0547-4. Respir Res. 2017. PMID: 28407775 Free PMC article.
-
The short and long telomere syndromes: paired paradigms for molecular medicine.Curr Opin Genet Dev. 2015 Aug;33:1-9. doi: 10.1016/j.gde.2015.06.004. Epub 2015 Jul 29. Curr Opin Genet Dev. 2015. PMID: 26232116 Free PMC article. Review.
-
Telomeres and age-related disease: how telomere biology informs clinical paradigms.J Clin Invest. 2013 Mar;123(3):996-1002. doi: 10.1172/JCI66370. Epub 2013 Mar 1. J Clin Invest. 2013. PMID: 23454763 Free PMC article. Review.
Cited by
-
High-throughput single telomere analysis using DNA microarray and fluorescent in situ hybridization.Nucleic Acids Res. 2024 Oct 28;52(19):e96. doi: 10.1093/nar/gkae812. Nucleic Acids Res. 2024. PMID: 39291738 Free PMC article.
-
Regulation of cellular senescence in tumor progression and therapeutic targeting: mechanisms and pathways.Mol Cancer. 2025 Apr 2;24(1):106. doi: 10.1186/s12943-025-02284-z. Mol Cancer. 2025. PMID: 40170077 Free PMC article. Review.
-
Telomeres in Space.Aging Cell. 2025 Mar;24(3):e70030. doi: 10.1111/acel.70030. Epub 2025 Mar 1. Aging Cell. 2025. PMID: 40022541 Free PMC article.
-
NANOME: A Nextflow pipeline for haplotype-aware allele-specific consensus DNA methylation detection by nanopore long-read sequencing.bioRxiv [Preprint]. 2025 Jul 4:2025.06.29.662079. doi: 10.1101/2025.06.29.662079. bioRxiv. 2025. PMID: 40631091 Free PMC article. Preprint.
-
Telomere-to-telomere DNA replication timing profiling using single-molecule sequencing with Nanotiming.Nat Commun. 2025 Jan 2;16(1):242. doi: 10.1038/s41467-024-55520-3. Nat Commun. 2025. PMID: 39747057 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

