Plasma proteomics identify biomarkers predicting Parkinson's disease up to 7 years before symptom onset
- PMID: 38890280
- PMCID: PMC11189460
- DOI: 10.1038/s41467-024-48961-3
Plasma proteomics identify biomarkers predicting Parkinson's disease up to 7 years before symptom onset
Abstract
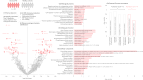
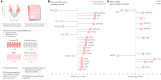
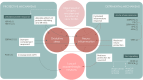
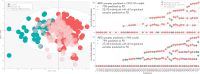
Parkinson's disease is increasingly prevalent. It progresses from the pre-motor stage (characterised by non-motor symptoms like REM sleep behaviour disorder), to the disabling motor stage. We need objective biomarkers for early/pre-motor disease stages to be able to intervene and slow the underlying neurodegenerative process. Here, we validate a targeted multiplexed mass spectrometry assay for blood samples from recently diagnosed motor Parkinson's patients (n = 99), pre-motor individuals with isolated REM sleep behaviour disorder (two cohorts: n = 18 and n = 54 longitudinally), and healthy controls (n = 36). Our machine-learning model accurately identifies all Parkinson patients and classifies 79% of the pre-motor individuals up to 7 years before motor onset by analysing the expression of eight proteins-Granulin precursor, Mannan-binding-lectin-serine-peptidase-2, Endoplasmatic-reticulum-chaperone-BiP, Prostaglaindin-H2-D-isomaerase, Interceullular-adhesion-molecule-1, Complement C3, Dickkopf-WNT-signalling pathway-inhibitor-3, and Plasma-protease-C1-inhibitor. Many of these biomarkers correlate with symptom severity. This specific blood panel indicates molecular events in early stages and could help identify at-risk participants for clinical trials aimed at slowing/preventing motor Parkinson's disease.
© 2024. The Author(s).
Conflict of interest statement
JH, MD, MX, SW, KB, ME, PG, MGB, CP, KM, ID, WH, JS, HV and CF and have no competing interests to report. MB has received funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – 413,501,650. CT has received honoraria for consultancy from Roche, and honoraria for educational lectures from UCB, and has received research funding for the PPMI study from the Michael J. Fox Foundation and funding from the EU (Horizon 2020) and stipends from the (International Parkinson’s and Movement Disorder Society) IPMDS. BM has received honoraria for consultancy from Roche, Biogen, AbbVie, UCB, and Sun Pharma Advanced Research Company. BM is a member of the executive steering committee of the Parkinson Progression Marker Initiative and PI of the Systemic Synuclein Sampling Study of the Michael J. Fox Foundation for Parkinson’s Research and has received research funding from the Deutsche Forschungsgemeinschaft (DFG), EU (Horizon 2020), Parkinson Fonds Deutschland, Deutsche Parkinson Vereinigung, Parkinson’s Foundation and the Michael J. Fox Foundation for Parkinson’s Research. MLM has received honoraria for speaking engagements from Deutsche Parkinson Gesellschaft e.V., and royalties from Gesellschaft fur Medien + Kommunikation mbH + Co. FSD has received honoraria for speaking engagements from AbbVie, Bial, Ever Pharma, Medtronic and royalties from Elsevier and Springer. She served on an advisory board for Zambon and Stada Pharma. FSD participated in Ad Boards for consultation: Abbvie, UCB, Bial, Ono, Roche and got honorary for lecturing: Stada Pharm, AbbVie, Alexion, Bial. S. Schade received institutional salaries supported by the EU Horizon 2020 research and innovation programme under grant agreement No. 863664 and by the Michael J. Fox Foundation for Parkinson’s Research under grant agreement No. MJFF-021923. He is supported by a PPMI Early Stage Investigators Funding Programme fellowship of the Michael J. Fox Foundation for Parkinson’s Research under grant agreement No. MJFF-022656. S. Schreglmann received institutional salaries supported by the EU Horizon 2020 research and innovation programme under grant agreement No. 863664, support from the Advanced Clinician Scientist programme by the Interdisciplinary Centre for Clinical Research, Wuerzburg, Germany, and from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Project-ID 424778381-TRR 295. He is a fellow of the Thiemann Foundation. He serves as a scientific adviser to Elemind Inc.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

