The miR-29 family facilitates the activation of NK-cell immune responses by targeting the B7-H3 immune checkpoint in neuroblastoma
- PMID: 38890285
- PMCID: PMC11189583
- DOI: 10.1038/s41419-024-06791-7
The miR-29 family facilitates the activation of NK-cell immune responses by targeting the B7-H3 immune checkpoint in neuroblastoma
Abstract
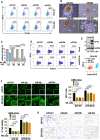
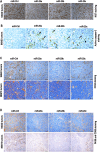
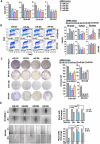
Neuroblastoma (NB) is a highly aggressive pediatric cancer that originates from immature nerve cells, presenting significant treatment challenges due to therapy resistance. Despite intensive treatment, approximately 50% of high-risk NB cases exhibit therapy resistance or experience relapse, resulting in poor outcomes often associated with tumor immune evasion. B7-H3 is an immune checkpoint protein known to inhibit immune responses. MicroRNAs (miRNAs) are small non-coding RNAs involved in post-transcriptional gene regulation. Our study aims to explore the impact of miRNAs on B7-H3 regulation, the anti-tumor immune response, and tumorigenicity in NB. Analysis of NB patients and patient-derived xenograft tumors revealed a correlation between higher B7-H3 expression and poorer patient survival. Notably, deceased patients exhibited a depletion of miR-29 family members (miR-29a, miR-29b, and miR-29c), which displayed an inverse association with B7-H3 expression in NB patients. Overexpression and knockdown experiments demonstrated that these miRNAs degrade B7-H3 mRNA, resulting in enhanced NK cell activation and cytotoxicity. In vivo, experiments provided further evidence that miR-29 family members reduce tumorigenicity, macrophage infiltration, and microvessel density, promote infiltration and activation of NK cells, and induce tumor cell apoptosis. These findings offer a rationale for developing more effective combination treatments that leverage miRNAs to target B7-H3 in NB patients.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Novel trispecific killer engager targeting B7-H3 enhances natural killer cell antitumor activity against head and neck cancer.J Immunother Cancer. 2025 Jul 23;13(7):e011370. doi: 10.1136/jitc-2024-011370. J Immunother Cancer. 2025. PMID: 40707133 Free PMC article.
-
A fc-engineered NKG2D × B7-H3 bispecific antibody enhances the antitumor activity by orchestrating cytotoxic lymphocytes.Int Immunopharmacol. 2025 Aug 28;161:115032. doi: 10.1016/j.intimp.2025.115032. Epub 2025 Jun 13. Int Immunopharmacol. 2025. PMID: 40516256
-
B7-H3 and CSPG4 co-targeting as Pan-CAR-T cell treatment of triple-negative breast cancer.J Immunother Cancer. 2025 May 26;13(5):e011533. doi: 10.1136/jitc-2025-011533. J Immunother Cancer. 2025. PMID: 40425233 Free PMC article.
-
Research progress of B7-H3 in malignant tumors.Front Immunol. 2025 May 30;16:1586759. doi: 10.3389/fimmu.2025.1586759. eCollection 2025. Front Immunol. 2025. PMID: 40519928 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
Cited by
-
Non-Coding RNAs and Innate Immune Responses in Cancer.Biomedicines. 2024 Sep 11;12(9):2072. doi: 10.3390/biomedicines12092072. Biomedicines. 2024. PMID: 39335585 Free PMC article. Review.
-
B7-H3 in Cancer Immunotherapy-Prospects and Challenges: A Review of the Literature.Cells. 2025 Aug 6;14(15):1209. doi: 10.3390/cells14151209. Cells. 2025. PMID: 40801642 Free PMC article. Review.
-
B7-H3 in glioblastoma and beyond: significance and therapeutic strategies.Front Immunol. 2024 Nov 25;15:1495283. doi: 10.3389/fimmu.2024.1495283. eCollection 2024. Front Immunol. 2024. PMID: 39664380 Free PMC article. Review.
-
B7H3 in Gastrointestinal Tumors: Role in Immune Modulation and Cancer Progression: A Review of the Literature.Cells. 2025 Apr 2;14(7):530. doi: 10.3390/cells14070530. Cells. 2025. PMID: 40214484 Free PMC article. Review.
-
Emerging clinical and research approaches in targeted therapies for high-risk neuroblastoma.Front Oncol. 2025 Mar 4;15:1553511. doi: 10.3389/fonc.2025.1553511. eCollection 2025. Front Oncol. 2025. PMID: 40104501 Free PMC article. Review.
References
-
- London WB, Castel V, Monclair T, Ambros PF, Pearson AD, Cohn SL, et al. Clinical and biologic features predictive of survival after relapse of neuroblastoma: a report from the International Neuroblastoma Risk Group project. J Clin Oncol. 2011;29:3286–92. doi: 10.1200/JCO.2010.34.3392. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

