High resolution long-read telomere sequencing reveals dynamic mechanisms in aging and cancer
- PMID: 38890299
- PMCID: PMC11189484
- DOI: 10.1038/s41467-024-48917-7
High resolution long-read telomere sequencing reveals dynamic mechanisms in aging and cancer
Abstract
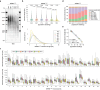
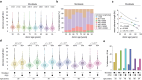
Telomeres are the protective nucleoprotein structures at the end of linear eukaryotic chromosomes. Telomeres' repetitive nature and length have traditionally challenged the precise assessment of the composition and length of individual human telomeres. Here, we present Telo-seq to resolve bulk, chromosome arm-specific and allele-specific human telomere lengths using Oxford Nanopore Technologies' native long-read sequencing. Telo-seq resolves telomere shortening in five population doubling increments and reveals intrasample, chromosome arm-specific, allele-specific telomere length heterogeneity. Telo-seq can reliably discriminate between telomerase- and ALT-positive cancer cell lines. Thus, Telo-seq is a tool to study telomere biology during development, aging, and cancer at unprecedented resolution.
© 2024. The Author(s).
Conflict of interest statement
C.H., J.R.J., K.A.F., F.H.G., and J.K. declare that they have no competing interests. T.T.S. has received travel funds to speak at events hosted by Oxford Nanopore Technologies. C.T., P.R., X.D., S.J., and S.H. are employees of Oxford Nanopore Technologies, Inc. and are stock or stock option holders in Oxford Nanopore Technologies plc. C.T., P.R., X.D., and S.H. are named as inventors on a patent application covering aspects of this work filed by Oxford Nanopore Technologies plc. Oxford Nanopore Technologies products are not intended for use for health assessment or to diagnose, treat, mitigate, cure, or prevent any disease or condition.
Figures



References
MeSH terms
Substances
Grants and funding
- GM142173/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 CA227934/CA/NCI NIH HHS/United States
- R01 CA234047/CA/NCI NIH HHS/United States
- AG0773424/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- CA234047/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 AG077324/AG/NIA NIH HHS/United States
- CA227934/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P30 CA014195/CA/NCI NIH HHS/United States
- R01 GM142173/GM/NIGMS NIH HHS/United States
- 19PABH134610000/American Heart Association (American Heart Association, Inc.)
- P30 AG062429/AG/NIA NIH HHS/United States
- ALTF 668-2019/European Molecular Biology Organization (EMBO)
- P30CA014195/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P30 AG068635/AG/NIA NIH HHS/United States

