AURKB/CDC37 complex promotes clear cell renal cell carcinoma progression via phosphorylating MYC and constituting an AURKB/E2F1-positive feedforward loop
- PMID: 38890303
- PMCID: PMC11189524
- DOI: 10.1038/s41419-024-06827-y
AURKB/CDC37 complex promotes clear cell renal cell carcinoma progression via phosphorylating MYC and constituting an AURKB/E2F1-positive feedforward loop
Abstract
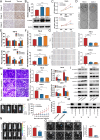
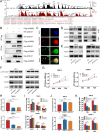
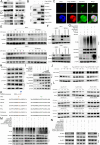
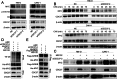
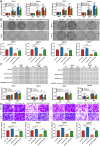
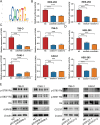
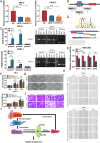
As the second most common malignant tumor in the urinary system, renal cell carcinoma (RCC) is imperative to explore its early diagnostic markers and therapeutic targets. Numerous studies have shown that AURKB promotes tumor development by phosphorylating downstream substrates. However, the functional effects and regulatory mechanisms of AURKB on clear cell renal cell carcinoma (ccRCC) progression remain largely unknown. In the current study, we identified AURKB as a novel key gene in ccRCC progression based on bioinformatics analysis. Meanwhile, we observed that AURKB was highly expressed in ccRCC tissue and cell lines and knockdown AURKB in ccRCC cells inhibit cell proliferation and migration in vitro and in vivo. Identified CDC37 as a kinase molecular chaperone for AURKB, which phenocopy AURKB in ccRCC. AURKB/CDC37 complex mediate the stabilization of MYC protein by directly phosphorylating MYC at S67 and S373 to promote ccRCC development. At the same time, we demonstrated that the AURKB/CDC37 complex activates MYC to transcribe CCND1, enhances Rb phosphorylation, and promotes E2F1 release, which in turn activates AURKB transcription and forms a positive feedforward loop in ccRCC. Collectively, our study identified AURKB as a novel marker of ccRCC, revealed a new mechanism by which the AURKB/CDC37 complex promotes ccRCC by directly phosphorylating MYC to enhance its stability, and first proposed AURKB/E2F1-positive feedforward loop, highlighting AURKB may be a promising therapeutic target for ccRCC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
NEIL3 promotes cell proliferation of ccRCC via the cyclin D1-Rb-E2F1 feedback loop regulation.DNA Repair (Amst). 2024 Jan;133:103604. doi: 10.1016/j.dnarep.2023.103604. Epub 2023 Nov 19. DNA Repair (Amst). 2024. PMID: 37992567
-
TRIP13-induced NUSAP1 upregulation promotes CcRCC progression through EMT and PI3K/AKT/mTOR pathway.J Transl Med. 2025 Aug 11;23(1):890. doi: 10.1186/s12967-025-06761-3. J Transl Med. 2025. PMID: 40790482 Free PMC article.
-
Biological and metabolomic insights into RACGAP1-mediated growth and progression of clear cell renal cell carcinoma.Am J Physiol Cell Physiol. 2025 Jul 1;329(1):C283-C297. doi: 10.1152/ajpcell.00066.2025. Epub 2025 Jun 16. Am J Physiol Cell Physiol. 2025. PMID: 40522863
-
A Systematic Review and Meta-analysis Comparing the Effectiveness and Adverse Effects of Different Systemic Treatments for Non-clear Cell Renal Cell Carcinoma.Eur Urol. 2017 Mar;71(3):426-436. doi: 10.1016/j.eururo.2016.11.020. Epub 2016 Dec 8. Eur Urol. 2017. PMID: 27939075
-
CYP1B1 promotes angiogenesis and sunitinib resistance in clear cell renal cell carcinoma via USP5-mediated HIF2α deubiquitination.Neoplasia. 2025 Aug;66:101186. doi: 10.1016/j.neo.2025.101186. Epub 2025 May 27. Neoplasia. 2025. PMID: 40435846 Free PMC article. Review.
Cited by
-
Histone lactylation-boosted AURKB facilitates colorectal cancer progression by inhibiting HNRNPM-mediated PSAT1 mRNA degradation.J Exp Clin Cancer Res. 2025 Aug 11;44(1):233. doi: 10.1186/s13046-025-03498-1. J Exp Clin Cancer Res. 2025. PMID: 40784984 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

