Tumor-resident microbiota contributes to colorectal cancer liver metastasis by lactylation and immune modulation
- PMID: 38890429
- PMCID: PMC11281901
- DOI: 10.1038/s41388-024-03080-7
Tumor-resident microbiota contributes to colorectal cancer liver metastasis by lactylation and immune modulation
Erratum in
-
Correction: Tumor-resident microbiota contributes to colorectal cancer liver metastasis by lactylation and immune modulation.Oncogene. 2025 Jul;44(24):2004-2007. doi: 10.1038/s41388-025-03439-4. Oncogene. 2025. PMID: 40456867 Free PMC article. No abstract available.
Abstract
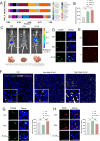
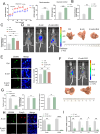
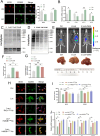
The role of tumor-resident microbiota in modulating tumor immunity remains unclear. Here, we discovered an abundance of intra-tumoral bacteria, such us E.coli, residing and resulting in Colorectal cancer liver metastasis (CRLM). E.coli enhanced lactate production, which mediated M2 macrophage polarization by suppressing nuclear factor-κB -gene binding (NF-κB) signaling through retinoic acid-inducible gene 1 (RIG-I) lactylation. Lactylation of RIG-I suppressed recruitment of NF-κB to the Nlrp3 promoter in macrophages, thereby reducing its transcription. This loss of Nlrp3 affected the immunosuppressive activities of regulatory T cells (Tregs) and the antitumor activities of and CD8+ T cells. Small-molecule compound screening identified a RIG-I lactylation inhibitor that suppressed M2 polarization and sensitized CRLM to 5-fluorouracil (5-FU). Our findings suggest that tumor-resident microbiota may be a potential target for preventing and treating CRLM.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

