Analysis of 10,478 cancer genomes identifies candidate driver genes and opportunities for precision oncology
- PMID: 38890488
- PMCID: PMC11387197
- DOI: 10.1038/s41588-024-01785-9
Analysis of 10,478 cancer genomes identifies candidate driver genes and opportunities for precision oncology
Abstract
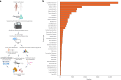
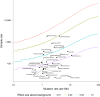
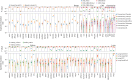
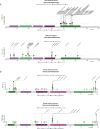
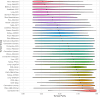
Tumor genomic profiling is increasingly seen as a prerequisite to guide the treatment of patients with cancer. To explore the value of whole-genome sequencing (WGS) in broadening the scope of cancers potentially amenable to a precision therapy, we analysed whole-genome sequencing data on 10,478 patients spanning 35 cancer types recruited to the UK 100,000 Genomes Project. We identified 330 candidate driver genes, including 74 that are new to any cancer. We estimate that approximately 55% of patients studied harbor at least one clinically relevant mutation, predicting either sensitivity or resistance to certain treatments or clinical trial eligibility. By performing computational chemogenomic analysis of cancer mutations we identify additional targets for compounds that represent attractive candidates for future clinical trials. This study represents one of the most comprehensive efforts thus far to identify cancer driver genes in the real world setting and assess their impact on informing precision oncology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
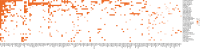







References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

