DeepLeish: a deep learning based support system for the detection of Leishmaniasis parasite from Giemsa-stained microscope images
- PMID: 38890604
- PMCID: PMC11186139
- DOI: 10.1186/s12880-024-01333-1
DeepLeish: a deep learning based support system for the detection of Leishmaniasis parasite from Giemsa-stained microscope images
Abstract
Background: Leishmaniasis is a vector-born neglected parasitic disease belonging to the genus Leishmania. Out of the 30 Leishmania species, 21 species cause human infection that affect the skin and the internal organs. Around, 700,000 to 1,000,000 of the newly infected cases and 26,000 to 65,000 deaths are reported worldwide annually. The disease exhibits three clinical presentations, namely, the cutaneous, muco-cutaneous and visceral Leishmaniasis which affects the skin, mucosal membrane and the internal organs, respectively. The relapsing behavior of the disease limits its diagnosis and treatment efficiency. The common diagnostic approaches follow subjective, error-prone, repetitive processes. Despite, an ever pressing need for an accurate detection of Leishmaniasis, the research conducted so far is scarce. In this regard, the main aim of the current research is to develop an artificial intelligence based detection tool for the Leishmaniasis from the Geimsa-stained microscopic images using deep learning method.
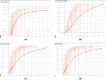
Methods: Stained microscopic images were acquired locally and labeled by experts. The images were augmented using different methods to prevent overfitting and improve the generalizability of the system. Fine-tuned Faster RCNN, SSD, and YOLOV5 models were used for object detection. Mean average precision (MAP), precision, and Recall were calculated to evaluate and compare the performance of the models.
Results: The fine-tuned YOLOV5 outperformed the other models such as Faster RCNN and SSD, with the MAP scores, of 73%, 54% and 57%, respectively.
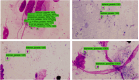
Conclusion: The currently developed YOLOV5 model can be tested in the clinics to assist the laboratorists in diagnosing Leishmaniasis from the microscopic images. Particularly, in low-resourced healthcare facilities, with fewer qualified medical professionals or hematologists, our AI support system can assist in reducing the diagnosing time, workload, and misdiagnosis. Furthermore, the dataset collected by us will be shared with other researchers who seek to improve upon the detection system of the parasite. The current model detects the parasites even in the presence of the monocyte cells, but sometimes, the accuracy decreases due to the differences in the sizes of the parasite cells alongside the blood cells. The incorporation of cascaded networks in future and the quantification of the parasite load, shall overcome the limitations of the currently developed system.
Keywords: Deep learning; Faster RCNN; Leishmaniasis; Microscopic image; Object detection; SSD; YOLOV5.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures

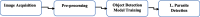



References
-
- von Chamier L, Laine RF, Henriques R. Artificial intelligence for microscopy: what you should know, Biochem. Soc. Trans 47;4:1029–1040, 2019, 10.1042/BST20180391. - PubMed
-
- Yu K-H, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2(10):719–31. 10.1038/s41551-018-0305-z. - PubMed
-
- Xing F, Yang L. Chapter 4 - Machine learning and its application in microscopic image analysis, in The Elsevier and MICCAI Society Book Series, G. Wu, D. Shen, and M. R. B. T.-M. L. and M. I. Sabuncu, Eds. Academic Press, 2016. 97–127.
-
- Mirbabaie M, Stieglitz S, Frick NRJ. Artificial intelligence in disease diagnostics: a critical review and classification on the current state of research guiding future direction. Health Technol (Berl) 2021;11(4):693–731. doi: 10.1007/s12553-021-00555-5. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

