Identification of Various Recombinants in a Patient Coinfected With the Different SARS-CoV-2 Variants
- PMID: 38890805
- PMCID: PMC11187932
- DOI: 10.1111/irv.13340
Identification of Various Recombinants in a Patient Coinfected With the Different SARS-CoV-2 Variants
Abstract
Background: Viral recombination that occurs by exchanging genetic materials between two viral genomes coinfecting the same host cells is associated with the emergence of new viruses with different virulence. Herein, we detected a patient coinfected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Delta and Omicron variants and identified various recombinants in the SARS-CoV-2 full-length spike gene using long-read and Sanger sequencing.
Methods: Samples from five patients in Japan with household transmission of coronavirus disease 2019 (COVID-19) were analyzed using molecular assays for detection and identification of SARS-CoV-2. Whole-genome sequencing was conducted using multiplex PCR with short-read sequencing.
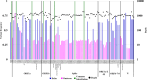
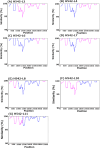
Results: Among the five SARS-CoV-2-positive patients, the mutation-specific assay identified the Delta variant in three, the Omicron variant in one, and an undetermined in one. The undermined patient was identified as Delta using whole-genome sequencing, but samples showed a mixed population of Delta and Omicron variants. This patient was analyzed for viral quasispecies by long-read and Sanger sequencing using a full-length spike gene amplicon. In addition to the Delta and Omicron sequences, the viral quasispecies analysis identified nine different genetic recombinant sequences with various breakpoints between Delta and Omicron sequences. The nine detected recombinant sequences in the spike gene showed over 99% identity with viruses that were detected during the Delta and Omicron cocirculation period from the United States and Europe.
Conclusions: This study demonstrates that patients coinfected with different SARS-CoV-2 variants can generate various viral recombinants and that various recombinant viruses may be produced during the cocirculation of different variants.
Keywords: SARS‐CoV‐2; coinfection; recombination; spike gene; viral quasispecies.
© 2024 The Author(s). Influenza and Other Respiratory Viruses published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

Similar articles
-
Sustained applicability of SARS-CoV-2 variants identification by Sanger Sequencing Strategy on emerging various SARS-CoV-2 Omicron variants in Hiroshima, Japan.BMC Genomics. 2024 Nov 11;25(1):1063. doi: 10.1186/s12864-024-10973-0. BMC Genomics. 2024. PMID: 39528931 Free PMC article.
-
Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events.Signal Transduct Target Ther. 2022 Apr 26;7(1):138. doi: 10.1038/s41392-022-00992-2. Signal Transduct Target Ther. 2022. PMID: 35474215 Free PMC article.
-
Evidence for SARS-CoV-2 Delta and Omicron co-infections and recombination.Med. 2022 Dec 9;3(12):848-859.e4. doi: 10.1016/j.medj.2022.10.002. Epub 2022 Oct 20. Med. 2022. PMID: 36332633 Free PMC article.
-
Structural impact of a new spike Y170W mutation detected in early emerging SARS-CoV-2 Omicron variants in France.Virus Res. 2024 May;343:199354. doi: 10.1016/j.virusres.2024.199354. Epub 2024 Mar 16. Virus Res. 2024. PMID: 38492859 Free PMC article. Review.
-
Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?J Virol. 2022 Mar 23;96(6):e0207721. doi: 10.1128/jvi.02077-21. Epub 2022 Mar 23. J Virol. 2022. PMID: 35225672 Free PMC article. Review.
Cited by
-
SARS-CoV-2 biological clones are genetically heterogeneous and include clade-discordant residues.J Virol. 2025 May 20;99(5):e0225024. doi: 10.1128/jvi.02250-24. Epub 2025 Apr 24. J Virol. 2025. PMID: 40272156 Free PMC article.
-
A general and biomedical perspective of viral quasispecies.RNA. 2025 Feb 19;31(3):429-443. doi: 10.1261/rna.080280.124. RNA. 2025. PMID: 39689947 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

