Characterisation and Expression Analysis of LdSERK1, a Somatic Embryogenesis Gene in Lilium davidii var. unicolor
- PMID: 38891306
- PMCID: PMC11174594
- DOI: 10.3390/plants13111495
Characterisation and Expression Analysis of LdSERK1, a Somatic Embryogenesis Gene in Lilium davidii var. unicolor
Abstract
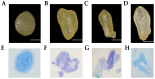
The Lanzhou lily (Lilium davidii var. unicolor) is a variant of the Sichuan lily of the lily family and is a unique Chinese 'medicinal and food' sweet lily. Somatic cell embryogenesis of Lilium has played an important role in providing technical support for germplasm conservation, bulb propagation and improvement of genetic traits. Somatic embryogenesis receptor-like kinases (SERKs) are widely distributed in plants and have been shown to play multiple roles in plant life, including growth and development, somatic embryogenesis and hormone induction. Integrating the results of KEGG enrichment, GO annotation and gene expression analysis, a lily LdSERK1 gene was cloned. The full-length open reading frame of LdSERK1 was 1875 bp, encoding 624 amino acids. The results of the phylogenetic tree analysis showed that LdSERK1 was highly similar to rice, maize and other plant SERKs. The results of the subcellular localisation in the onion epidermis suggested that the LdSERK1 protein was localised at the cell membrane. Secondly, we established the virus-induced gene-silencing (VIGS) system in lily scales, and the results of LdSERK1 silencing by Tobacco rattle virus (TRV) showed that, with the down-regulation of LdSERK1 expression, the occurrence of somatic embryogenesis and callus tissue induction in scales was significantly reduced. Finally, molecular assays from overexpression of the LdSERK1 gene in Arabidopsis showed that LdSERK1 expression was significantly enhanced in the three transgenic lines compared to the wild type, and that the probability of inducing callus tissue in seed was significantly higher than that of the wild type at a concentration of 2 mg/L 2,4-D, which was manifested by an increase in the granularity of the callus tissue.
Keywords: LdSERK1; Lilium davidii var. unicolor; gene isolation; phenotyping; somatic embryogenesis receptor-like kinase (SERK) gene.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

