Genome-Wide Association Study Identifies Quantitative Trait Loci and Candidate Genes Involved in Deep-Sowing Tolerance in Maize (Zea mays L.)
- PMID: 38891341
- PMCID: PMC11175157
- DOI: 10.3390/plants13111533
Genome-Wide Association Study Identifies Quantitative Trait Loci and Candidate Genes Involved in Deep-Sowing Tolerance in Maize (Zea mays L.)
Abstract
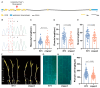
Deep sowing is an efficient strategy for maize to ensure the seedling emergence rate under adverse conditions such as drought or low temperatures. However, the genetic basis of deep-sowing tolerance-related traits in maize remains largely unknown. In this study, we performed a genome-wide association study on traits related to deep-sowing tolerance, including mesocotyl length (ML), coleoptile length (CL), plumule length (PL), shoot length (SL), and primary root length (PRL), using 255 maize inbred lines grown in three different environments. We identified 23, 6, 4, and 4 quantitative trait loci (QTLs) associated with ML, CL, PL, and SL, respectively. By analyzing candidate genes within these QTLs, we found a γ-tubulin-containing complex protein, ZmGCP2, which was significantly associated with ML, PL, and SL. Loss of function of ZmGCP2 resulted in decreased PL, possibly by affecting the cell elongation, thus affecting SL. Additionally, we identified superior haplotypes and allelic variations of ZmGCP2 with a longer PL and SL, which may be useful for breeding varieties with deep-sowing tolerance to improve maize cultivation.
Keywords: ZmGCP2; deep sowing; genome-wide association study; maize; plumule length; quantitative trait locus; shoot length.
Conflict of interest statement
Author Kun Hu is employed by the company “Sinograin Chengdu Storage Research Institute Co., Ltd., Chengdu, China”. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Hubert B., Rosegrant M., van Boekel M.A., Ortiz R. The Future of Food: Scenarios for 2050. Crop Sci. 2010;50:33–50. doi: 10.2135/cropsci2009.09.0530. - DOI
-
- Wang Y.Z., Liu Z.Q., Wang G.Q., Li Y. Investigation on Relationship between High Yield and Population’s Uniformity in Maize. J. Maize Sci. 2000;8:43–45.
-
- Feng P., Wang B., Harrison M.T., Wang J., Liu K., Huang M., Liu D.L., Yu Q., Hu K. Soil Properties Resulting in Superior Maize Yields upon Climate Warming. Agron. Sustain. Dev. 2022;42:85. doi: 10.1007/s13593-022-00818-z. - DOI
-
- Du L., Jiang H., Zhao G., Ren J. Gene Cloning of ZmMYB59 Transcription Factor in Maize and Its Expression during Seed Germination in Response to Deep-sowing and Exogenous Hormones. Plant Breed. 2017;136:834–844. doi: 10.1111/pbr.12550. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

