Genome Sequence Comparisons between Small and Large Colony Phenotypes of Equine Clinical Isolates of Arcanobacterium hippocoleae
- PMID: 38891657
- PMCID: PMC11171008
- DOI: 10.3390/ani14111609
Genome Sequence Comparisons between Small and Large Colony Phenotypes of Equine Clinical Isolates of Arcanobacterium hippocoleae
Abstract
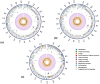
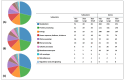
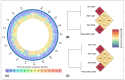
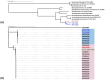
Arcanobacterium hippocoleae is a Gram-positive fastidious bacterium and is occasionally isolated from the reproductive tract of apparently healthy mares (Equus caballus) or from mares with reproductive tract abnormalities. Apart from a few 16S rRNA gene-based GenBank sequences and one recent report on complete genome assembly, detailed genomic sequence and clinical experimental data are not available on the bacterium. Recently, we observed an unusual increase in the detection of the organism from samples associated with mare reproductive failures in Atlantic Canada. Two colony morphotypes (i.e., small, and large) were detected in culture media, which were identified as A. hippocoleae by MALDI-TOF mass spectrometry and 16S rRNA gene sequencing. Here, we report the whole genome sequencing and characterization of the morphotype variants. The genome length of the large phenotypes was between 2.42 and 2.43, and the small phenotype was 1.99 Mbs. The orthologous nucleotide identity between the large colony phenotypes was ~99%, and the large and small colony phenotypes was between 77.86 and 78.52%, which may warrant the classification of the two morphotypes into different species. Phylogenetic analysis based on 16S rRNA genes or concatenated housekeeping genes grouped the small and large colony variants into two different genotypic clusters. The UvrA protein, which is part of the nucleotide excision repair (NER) system, and 3-isopropoylmalate dehydratase small subunit protein expressed by the leuD gene were identified as potential virulence factors in the large and small colony morphotypes, respectively. However, detailed functional studies will be required to determine the exact roles of these and other identified hypothetical proteins in the cellular metabolism and potential pathogenicity of A. hippocoleae in mares.
Keywords: A. hippocoleae; NGS; Oxford Nanopore; mares; reproductive tract.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Isolation and comparison of Arcanobacterium hippocoleae isolates from the genital tract of 15 mares.Vet Microbiol. 2019 Jan;228:129-133. doi: 10.1016/j.vetmic.2018.11.026. Epub 2018 Nov 28. Vet Microbiol. 2019. PMID: 30593358
-
Identification of Arcanobacterium hippocoleae by MALDI-TOF MS analysis and by various genotypical properties.Res Vet Sci. 2017 Dec;115:10-12. doi: 10.1016/j.rvsc.2017.01.006. Epub 2017 Jan 17. Res Vet Sci. 2017. PMID: 28126698
-
Arcanobacterium hippocoleae sp. nov., from the vagina of a horse.Int J Syst Evol Microbiol. 2002 Mar;52(Pt 2):617-619. doi: 10.1099/00207713-52-2-617. Int J Syst Evol Microbiol. 2002. PMID: 11931175
-
Isolation of Arcanobacterium hippocoleae from a case of placentitis and stillbirth in a mare.J Vet Diagn Invest. 2008 Sep;20(5):688-91. doi: 10.1177/104063870802000532. J Vet Diagn Invest. 2008. PMID: 18776114
-
Multi-year comparison of VITEK® MS and 16S rRNA gene sequencing performance for the identification of rarely encountered anaerobes causing invasive human infections in a large Canadian region: can our laboratory abandon 16S rRNA gene sequencing?Anaerobe. 2022 Dec;78:102640. doi: 10.1016/j.anaerobe.2022.102640. Epub 2022 Sep 17. Anaerobe. 2022. PMID: 36126828
References
-
- Wickhorst J.P., Sammra O., Hassan A.A., Alssashen M., Lämmler C., Prenger-Berninghoff E., Erhard M., Metzner M., Paschertz K., Timke M., et al. Identification of Arcanobacterium hippocoleae by MALDI-TOF MS analysis and by various genotypical properties. Res. Vet. Sci. 2017;115:10–12. doi: 10.1016/j.rvsc.2017.01.006. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

