Cordycepin Triphosphate as a Potential Modulator of Cellular Plasticity in Cancer via cAMP-Dependent Pathways: An In Silico Approach
- PMID: 38891880
- PMCID: PMC11171877
- DOI: 10.3390/ijms25115692
Cordycepin Triphosphate as a Potential Modulator of Cellular Plasticity in Cancer via cAMP-Dependent Pathways: An In Silico Approach
Abstract
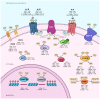
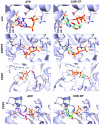
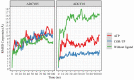
Cordycepin, or 3'-deoxyadenosine, is an adenosine analog with a broad spectrum of biological activity. The key structural difference between cordycepin and adenosine lies in the absence of a hydroxyl group at the 3' position of the ribose ring. Upon administration, cordycepin can undergo an enzymatic transformation in specific tissues, forming cordycepin triphosphate. In this study, we conducted a comprehensive analysis of the structural features of cordycepin and its derivatives, contrasting them with endogenous purine-based metabolites using chemoinformatics and bioinformatics tools in addition to molecular dynamics simulations. We tested the hypothesis that cordycepin triphosphate could bind to the active site of the adenylate cyclase enzyme. The outcomes of our molecular dynamics simulations revealed scores that are comparable to, and superior to, those of adenosine triphosphate (ATP), the endogenous ligand. This interaction could reduce the production of cyclic adenosine monophosphate (cAMP) by acting as a pseudo-ATP that lacks a hydroxyl group at the 3' position, essential to carry out nucleotide cyclization. We discuss the implications in the context of the plasticity of cancer and other cells within the tumor microenvironment, such as cancer-associated fibroblast, endothelial, and immune cells. This interaction could awaken antitumor immunity by preventing phenotypic changes in the immune cells driven by sustained cAMP signaling. The last could be an unreported molecular mechanism that helps to explain more details about cordycepin's mechanism of action.
Keywords: adenylate cyclase; cordycepin; mechanism of action; molecular docking; molecular dynamics; purine metabolites; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Cordycepin activates AMP-activated protein kinase (AMPK) via interaction with the γ1 subunit.J Cell Mol Med. 2014 Feb;18(2):293-304. doi: 10.1111/jcmm.12187. Epub 2013 Nov 28. J Cell Mol Med. 2014. PMID: 24286368 Free PMC article.
-
Cordycepin (3'-deoxyadenosine) inhibits human platelet aggregation in a cyclic AMP- and cyclic GMP-dependent manner.Eur J Pharmacol. 2007 Mar 8;558(1-3):43-51. doi: 10.1016/j.ejphar.2006.11.073. Epub 2006 Dec 12. Eur J Pharmacol. 2007. PMID: 17229422
-
A molecular dynamics study of adenylyl cyclase: The impact of ATP and G-protein binding.PLoS One. 2018 Apr 25;13(4):e0196207. doi: 10.1371/journal.pone.0196207. eCollection 2018. PLoS One. 2018. PMID: 29694437 Free PMC article.
-
Cordycepin for Health and Wellbeing: A Potent Bioactive Metabolite of an Entomopathogenic Cordyceps Medicinal Fungus and Its Nutraceutical and Therapeutic Potential.Molecules. 2020 Jun 12;25(12):2735. doi: 10.3390/molecules25122735. Molecules. 2020. PMID: 32545666 Free PMC article. Review.
-
Regulating cellular cyclic adenosine monophosphate: "Sources," "sinks," and now, "tunable valves".Wiley Interdiscip Rev Syst Biol Med. 2020 Sep;12(5):e1490. doi: 10.1002/wsbm.1490. Epub 2020 Apr 23. Wiley Interdiscip Rev Syst Biol Med. 2020. PMID: 32323924 Free PMC article. Review.
References
-
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

